
An interspecies pair of C. trifasiatus (left) and C. melapterus (right) seen at Socotra. Credit: Tane Sinclair-Taylor
In recent years, there has been a flurry of published research into the causes of speciation in coral reef fishes. For many Indo-Pacific groups, geography seems to be the main driver of diversity, but it isn’t always well-known how this regional diversity was formed and when this diversification occurred. Such answers are best sought by studying subtle differences in genetic sequences—the greater the molecular difference, the greater the age at which specimens last shared a common ancestor.
In a newly published article by Waldrop et al, these techniques were applied to a widespread and closely related subgenus of butterflyfish: Corallochaetodon. Four distinctive species make up this lineage, all occupying parapatric distributions spanning from the Red Sea to Polynesia. The names aren’t likely to be terribly familiar to the average reef aquarist, as these are all obligate corallivores (i.e. they only eat Acropora), but these are nonetheless some of the most ubiquitous members of their family to be found in healthy reef environments.
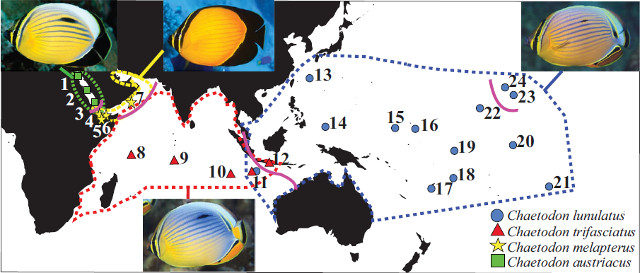
Biogeography of Chaetodon (Corallochaetodon) group. Credit: Walldrop et al 2016
Of the four, C. lunulatus of the Pacific Ocean and C. trifasciatus of the Indian Ocean are the two most widely distributed. This geminate species pair differs mostly in color, with trifasciatus having a bluer color to the sides and more orange along the caudal peduncle. The remaining species, C. austriacus of the Red Sea and C. melapterus of the Arabian Gulf, are dramatically more vibrant in color, having bright orange bodies and mostly black fins posteriorly. None of these are seen with any regularity in the aquarium trade, which is certainly in the best interests of these finicky fishes.
As butterflyfishes tend to have a relatively long-lasting pelagic larval stage to their development, it’s not unexpected that they should show fairly widespread populations. Relatively few members of the family show the kind of regionalized endemism seen in more sedentary groups like Amphiprion or Chrysiptera; instead, we see most chaetodontid species spanning entire ocean basins or oftentimes encompassing the full breadth of the tropical Indo-Pacific.

The Pacific Ocean C. lunulatus. Credit: Paddy Ryan
In analyzing the genetics of Corallochaetodon, Waldrop et al found support for the Indian and Pacific Ocean populations splitting approximately 3 MYA, roughly correlating with a period some 2.6-2.8 MYA when sea levels dropped and the Sunda Shelf of Indonesia effectively isolated these two basins and the fish populations therein. This is not a particularly novel finding, as similar results have been seen in many other sister species in this region, though this age is older by around one million years from that seen in other butterflyfishes studied to date.
Of more interest is the intraoceanic population data examined, which suggests significant population structure across the Pacific Ocean. This is the nitty gritty for how new species are formed, as regional differences in population genetics are the first step towards the formation of reproductively isolated lineages (i.e. species). In the Corallochaetodon study, specimens from Moorea in French Polynesia, from Fiji, and those from Hawaii showed particularly high levels of genetic divergence relative to those elsewhere in the Pacific. Hawaiian specimens even suggested the presence of a deep genetic divide between those from the main islands and those from the more remote reefs of the northwest. The authors state that the island of Kaua’i, which marks the boundary between these two populations, is mostly devoid of specimens, suggesting a mechanism serving to isolate the two.
In contrast to the cryptic genetic diversity seen in the Pacific, specimens from the Indian Ocean are apparently far more homogenous, with only those from the relatively obscure Chagos Archipelago (south of the Maldives) showing any real divergence. Unfortunately, specimens from the East African coastline and the Mascarenes were unavailable for inclusion in this study. These two regions tend to show a considerable amount of endemism relative to the rest of the Indian Ocean, but, alas, this Corallochaetodon query will be left unresolved for now.

The Red Sea endemic C. austriacus. Credit: Bernard E. Picton
When it comes to the more restricted fishes of the Red Sea and Arabian Peninsula, the two species present here, despite showing easily observable differences to each other, tend to share a common haplotype (i.e. a common genetic sequence). These two are, in turn, closest genetically to the Indian Ocean C. trifasciatus, differing in only 3 “diagnostic nucleotide substitutions” of their cytochrome b mitochondrial gene. Compare this to the 28 substitutions seen between the Indian and Pacific Ocean taxa and it becomes clear which speciation event occurred first.
The authors put an estimated divergence for the Red Sea and Arabian species from the more widespread Indian Ocean population at 0.75 MYA, a period during which the Red Sea was frequently cut off by lowered sea levels. On the other hand, the Red Sea C. austriacus and the Arabian C. melapterus seem to have split from each other as recently as 50,000 years ago, and, while the two do overlap to some small extent in the southernmost portions of the Red Sea, this only became possible in the last 20,000 years when the Red Sea regained its connection to the Indian Ocean. Even today, this area is a marginal habitat for reef fishes, with temperatures and salinities that can fluctuate dramatically.
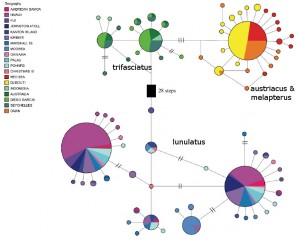
Haploytype map, with relative abundance of a haplotype indicated by the size of the circles. Note how diverse the Red Sea * Arabian species are compared to the more widespread Indian Ocean taxon. Modified from Walldrop et al 2016
Perhaps the most intriguing finding of this study was that the genetic diversity within these four species is surprisingly comparable. We find a similar number of distinct haplotypes in the Red Sea as we find in the more widespread Indian and Pacific Ocean populations, which is somewhat counterintuitive. The authors suggest that the abundance of C. austriacus and melapterus (recall that these are some of the most frequently encountered species of chaetodontids within their range) might explain their heterogenous genetic makeup, but this answer doesn’t seem all that compelling. Shouldn’t the same phenomenon be present for C. trifasciatus and lunulatus then… shouldn’t they show even more diversity given their much larger distributions?
There are obvious parallels that can be drawn from this study to many other groups of reef fishes with a similar distribution. Think of the recognized morphological differences between Indian and Pacific Ocean Imperator and Regal Angelfishes and it becomes clear how much work remains when it comes to reef fish taxonomy. With each new study into reef fish biogeography and population genetics we come even closer to cracking the code for how and why these creatures speciated so explosively.








0 Comments