
Currently unavailable obligate corallivore orangeface butterflyfish (Chaetodon larvatus) may have exhibited dietary plasticity during their putative migration through the Suez Canal to the Mediterranean (Adams 2018).
Identical processes underpin the wellbeing of the various microclimate niches (microbiomes) of organisms including corals and fish, which are economically important insofar as significant quantities of the fish consumed by humans are farmed and they can transmit zoonoses. Sea cages stress fish due to overcrowding, handling, and meaningfully polluting feeds, which leaves them susceptible to diseases, several of which have emerged over the last two decades (Fig 1.; Mougin & Joyce 2022) where wild and captive fish holobiont communities differ (Muñoz-Baquero et al. 2023). Antibiotic resistance has been acquired by many prokaryotic fish pathogens whereas several countries have banned their use and copper-based remedies for food fish. The industry has thus been forced to explore holistic methods of control which heighten natural defenses. Numerous studies are funded to investigate the mucosal microbiota of fish that inhabit their skin, gills, gut, nasal, and oral cavities which are primary routes of infection, while the environment rather than the host influences their community compositions (Lorgen-Ritchie et al. 2023). Cohorts of commensal and mutualistic microbes are central to the maintenance of an effective barrier, where destabilization and dysbiosis occur sometime before the onset of pathology. It is imperative therefore to identify biomarkers which assist early detection for timely disease-mitigating probiotic and prebiotic manipulations. If such therapies are effective in the over-disquieting milieus of mariculture, they cannot fail to enhance the vitality and survivorship of ornamental marine fish in optimized reefs or fish-only systems. Few grants are awarded to investigate fish destined for aquaria, so we necessarily simplify and present the findings of mariculture research, while the previous editorials in this series are considered foundational which support interpretation.
We cannot eradicate disease in reefs like we can in fish-only systems. Appreciation of holobiont and pathogen gene expression and the dynamics of their adaptive, biocidal, immunological, and virulence strategies remain central to the development of cures, which must respect the housekeeping commensals that become opportunistic pathogens when holobiont defenses are compromised or when fish integuments are breached. Cutting-edge perspectives have commenced to regard all microbes as opportunistic including Koch’s postulate-affirmed aetiological agents, while proposing disease originates from perturbations in microbial function which initiates homeostatic destabilization and diminished immunity (Mougin & Joyce 2022).

Fig 1. Mariculture sea cages raising seabass and sea bream off Galaxidi in Greece.
We are better placed to understand what constitutes a “healthy” population after studying the various microbial communities in the previous articles. We comprehend that diversity, richness, and evenness are vital, however from an objective standpoint it is challenging to define the exact composition that contributes to function. A cross-taxa core microbiome that supports finfish homeostasis is unlikely to exist (Lorgen-Ritchie et al. 2023). Instead Mougin and Joyce (2022) studied a specified pathogen and dysbiotic state which contributed to aetiology, diagnosis, and therapy. They theorized disease is proceeded by a loss of “healthy” prokaryotic phylotypes and their adaptive remedial competencies described as metagenomic plasticity. It is unclear if dysbiosis causes or is a symptom of disease, nevertheless three types are exemplified by a loss of beneficial microbes (i), constrained microbial diversity (ii), and the emergence of an aberrant holobiont (pathobiont; iii), all of which have discernible features (Mougin & Joyce 2022).
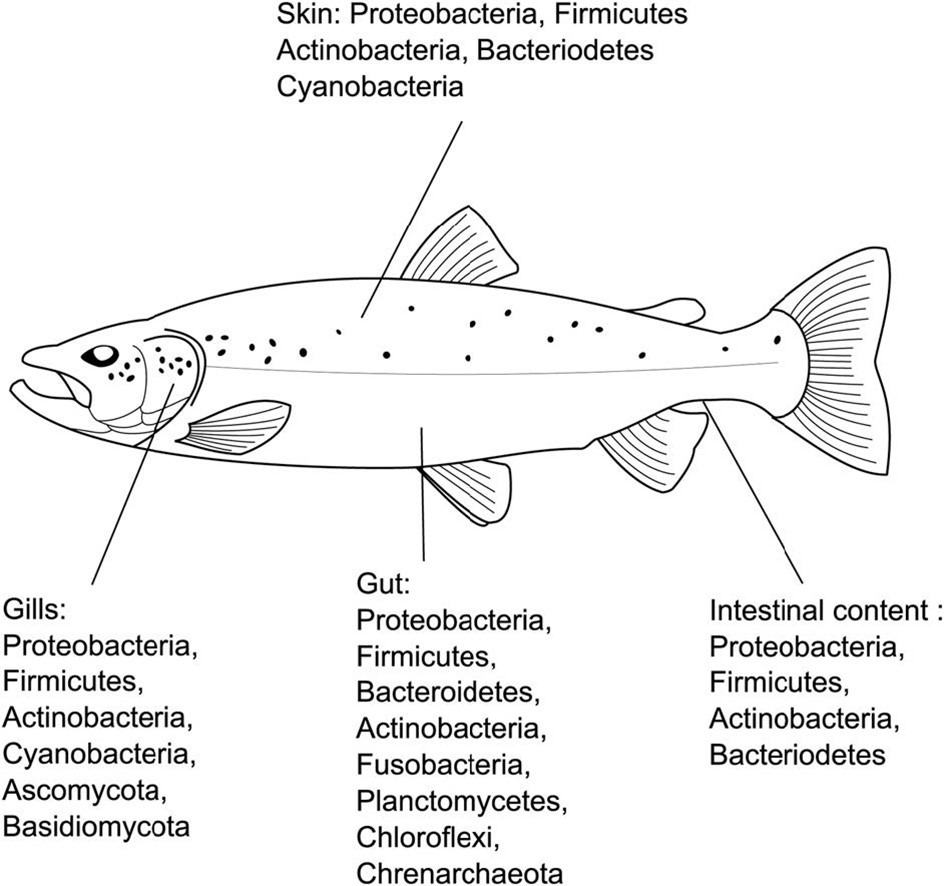
Around 90 percent of all seawater biomass is microscopic, and the microbiomes of its inhabitants are inextricably linked to the microbial profile of their surroundings (Edgerton et al. 2018). The gut, skin, and gills of fish have defined microbial communities yet more is known of the gut mucosa and its populations due to its importance in contagion and aetiology, and an endocrine, neuro-, and immunological association called the gut-brain axis (Rosado et al. 2022). Furthermore marine teleosts must continuously drink water to assist osmoregulation as all marine life must guard against desiccation, where pathology of the gut mucosae elicits “leaky” interstitial (intercellular) junctions (Sitjà-Bobadilla et al. 2019) which assist pathogen infiltration. Likewise the paracellular routes of marine fish gill are easier to negotiate compared with those of freshwater fish. Intestinal epithelial surfaces and their microbial consortia exhibit profound antimicrobial competences (Fouz et al. 2000) however some pathogens may be internalized by glycoprotein or actin-dependent endocytosis where they escape destruction (López-Dóriga et al. 2000; Andreoni & Magnani 2014).
It was concluded in the late 20th century that fish intestines harbored limited microflora, however less than 1 percent of microbes could be isolated and identified using the agar of the time (Amann et al. 1995) so gut mucosal microbiomes were grossly underestimated (Edgerton et al. 2018). Molecular techniques now indicate that such analysis could only detect 0.1 percent of fish gut affiliates (Zhou et al. 2014).
Numerous studies have recognized a significant perturbation of gut microbes during enteritis, although such changes are not localized because cross-holobiont communities may shift. Gut infections can transform the microbial communities of skin and gills (Legrand et al. 2018) albeit dynamic alterations in populations arise as aetiologies advance (Mougin & Joyce 2022).
The adaptive and innate immunity of higher eukaryotes will exert a more pronounced effect on the microbial communities of their microbiomes compared to corals with merely an innate response. All mucosal surfaces are flooded with compliment, oxidative bursts, antibodies, and numerous phagocytic lymphocytes.
Next-generation sequencing (NGS) techniques like the algorithm linear discriminant analysis effect size (LEfSe) identifies metagenomic biomarkers of key prokaryotic species, their gene expression and metabolism, and discriminates between microbial communities with unique profiles. That said, such technology has yet to be used in aquaculture (Segata et al. 2011, cited in Mougin & Joyce 2022). Multi“omic” approaches integrate taxonomic, metagenome, metatranscriptome, and metaproteome databases, with biomarkers of microbial networks, prokaryotic defense, and metabolism (Bass et al. 2019).
A loss of diversity in gut microflora is associated with overgrowth with a handful of species, however the phyla Fusobacteria and Firmicutes have been recognized as members of beneficial consortia. A study suggested a decline in these lineages and an increase of Proteobacteria was commensurate with disease (Miyake et al. 2020). Mycoplasma species (Firmicutes) have also been identified as a genus that is present in “healthy” populations which decline before the onset of pathology, however other studies have linked the genus Mycoplasma with pathogenesis. Cetobacterium species were found in healthy populations where they synthesise vitamin B12 and their decline was indicative of contagions, whereas proliferation of the genera Aeromonas, Shewanella, and Vibrio was redolent of pathology (Mougin & Joyce 2022).
Several studies have noticed a change in bacterial composition accompanies infections of the skin and gill. The prokaryotic genera Acinetobacter, Shewanella, and Pseudomonas were curtailed on the skin of freshwater trout during colonisation with “white spot” (Ichthyophthirius multifiliis; Zhang et al. 2018; Mougin & Joyce 2022). Changes in the biochemical profile of mucus and a declination of the genera Alteromonas, Thalassabius, and Winogradskyella and trivial blooms of Flavobacterium, Chryseobacterium, and Tenacibaculum species were associated with skin ulcerations of gilthead seabream (Sparus aurata; Tapia-Paniagua et al. 2018), whereas two further studies suggested mucosal Rubritalea species were integral to the healthy consortia of European seabass (Dicentrarchus labrax; Rosado et al. 2019a; Cámara-Ruiz et al. 2021) where the known pathogen Vibrio harveyi increased diversity (Cámara-Ruiz et al. 2021). Similarly, Stenotrophomonas, Polaribacter 4, Pseudomonas, and Rubritalea species were lost from the gills of Dicentrarchus labrax infected with Photobacterium damselae piscicida (Rosado et al. 2019a). Rubritalea species are core cross-anatomy affiliates of Dicentrarchus labrax and Sparus aurata (Rosado et al 2019b; Rosado et al. 2021) which provide carotenoids, squalene, other antioxidants and beneficial metabolites (Spanova & Daum 2011; Yoon et al. 2018). Nevertheless this genus is merely indicative of the beneficial consortia of these species (Mougin & Joyce 2022).
- Fig 2. Finfish microbiome-associated bacterial phyla and fish health dynamics.
- Images courtesy of Llewellyn et al. 2017 (left) and Mougin and Joyce 2022 and their Creative Commons Attribution Licences.
Oleispira species declined on the skin of Atlantic salmon (Salmo salar) infected with salmonid alphavirus (SAV) where these microbes somehow assist the saltwater acclimation (smoltification) of parr and appear to shield against pathogens in triploid fish with three copies of each chromosome (Reid et al. 2017; Brown et al. 2021) insofar as 3n is routinely induced in aquacultured salmonids to preclude wildtype crosses. The ratio of the phyla Proteobacteria to Bacteroidetes decreases in skin mucus with the onset of enteritis in yellowtail kingfish (Seriola lalandi) where such ratios maybe useful for early generic biomarkers where wild kingfish exhibit higher ratios (Legrand et al. 2019).
Proteobacterial proliferation in fish alimentary canals is a nonergonomic warning signature insofar as fish must be destroyed for analysis (Miyake et al. 2020) while Pseudomonas species have been associated with diseased Yunlong grouper (Epinephelus species; Ma et al. 2019). Waterborne Vibrionaceae correspond to populations inhabiting cultured fish, and thus the profile of planktonic bacteria appears a valuable barometer of livestock health (Kim & Lee 2017). Dysbiosis and disease has been associated with the opportunistic genera Chryseobacterium (Weeksellaceae), Flavobacterium (Flavobacteriaceae), Granulicatella (Carnobacteriaceae), Pseudomonas (Pseudomonadaceae), Streptococcus (Streptococcaceae), Tenacibaculum (Flavobacteriaceae), and Vibrio (Vibrionaceae; Llewellyn et al. 2017; Reid et al. 2017; Tapia-Paniagua et al. 2018; Zhang et al. 2018).
Quests for taxonomic biomarkers have failed to provide the necessary insight likely due to confounding serotypes, constantly evolving species and strains, species-specific interactions as well as abiotic and ecological factors (Mougin & Joyce 2022). Screening techniques must identify the entire profile of beneficial microbial communities under all types of environmental conditions for each species before such investigations yield useful findings. Too numerous to quantify variables thwart these investigations where ergonomic and efficacious molecular techniques are required.
Holistic approaches use high-throughput molecular screening to profile prokaryotic communities that propose populations are shaped by host immunity and gene expression (Wang & Loreau 2014; Kim et al. 2017) which is supported by our observations in the preceding editorials. Studies suggest that low diversity is nonindicative of dysbiosis and disease (Bozzi et al. 2021) because increments or decrements can head pathology onset (Bass et al. 2019; Mougin & Joyce 2022). Such findings redirect our attention to community shifts irrespective of prokaryotic phylotypes, diversity, evenness, and richness, while population profiles are restructured temporally by aetiology and defense (Mougin & Joyce 2022).
Functional redundancy and the identification of cross-population gene homologues or metabolic signatures within mucus may prove worthwhile biomarkers, where tools like PICRUSt cross-reference 16S rRNA databases with metagenomic function (Langille et al. 2013; Ortiz-Estrada et al. 2019). Such analysis has its limitations inasmuch as BLAST searches may unearth unexpressed/dormant genes or vestigial non-functional homologues or those that have evolved divergently to perform unpredictable roles, while databases may lack phylogenetic breadth and depth (Mougin & Joyce 2022).
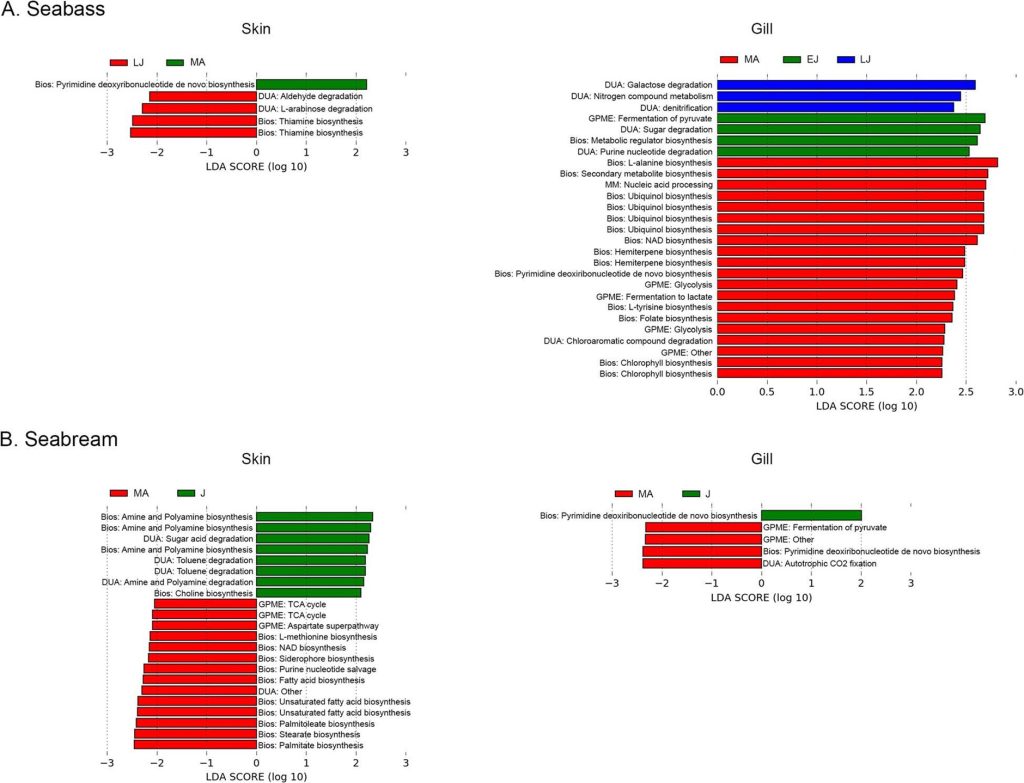
Fig 3. Skin and gill metabolome analyses of seabream and European Seabass: early juveniles [EJ]; late juveniles [LJ]; juveniles [J]; mature adults [MA]; biosynthesis [Bios]; degradation/utilization/assimilation [DUA]; generation of precursor metabolites and energy [GPME], and macromolecule modification [MM]. Analyses and image courtesy of Rosado et al. 2021 and the Creative Commons Attribution License.
Any method for monitoring maricultured specimens must be non-invasive, non-stressful and convenient, such as ventral or flank scrapes and/or water analysis, however the latter appears to promote inconsistency (Mougin & Joyce 2022). Perhaps investigations of water are only useful during an active infection with high planktonic loads.
We cannot make progress unless we unravel the mechanisms that underpin eubiosis and dysbiosis and ascertain which comes first: microbial community perturbations or disease (Mougin & Joyce 2022). Remarkably this area of research seems less explored than the microbiomes of corals and the holobiont network that supports and reinforces coral homeostasis and symbiosis. Nevertheless, what we have learnt from corals may be applicable.
Corals sift and selectively nurture residual symbionts and/or recruit them from the environment during abiotic disruptions in accordance with the microbial flexibility hypothesis. The prokaryotic holobiont communities of corals alter gene expression where metagenomic plasticity facilitates shifts in their population dynamics in the face of mild or moderate stress. Replete microbiomes demand extensive purging to accommodate recruitment and restructuring yet survival necessitates timely reversion. It is advised that dysbiosis is a natural and intended response for corals to a chronic stressor that exceeds the mitigating capacity of their microbes. Common symptoms of fish disease include surplus mucus and/or shedding of gut epithelium seen as a white stringy anal discharge. It is likely such responses are immunological and instigated by the host, like a raised temperature in human infections or coral bleaching, because mucosal sloughing also rids the fish of surface microbes. Signatures likely arise in fish mucus in response to pathogenic colonization, or after significant flushing to re-establish a healthy community, yet it may be too late to use them for early detection (Boilard et al. 2020; Mougin & Joyce 2022; Aslett 2024b).
Studies have focused on the bacterial affiliates of fish because they are likely candidates for mitigating illness and expediting recovery, however microeukaryotes and Archaea are present in fish holobionts that may manufacture detectable biomarkers. Viruses including bacteriophage are present which constitute the virome, which mobilize nucleic acid and may thus transform the virulence or environmental compliance of their lysogenic hosts. Alas, lytic bacteriophages mutate too frequently to be used as therapies to destroy pathogens, yet they may be engineered while they retain an aptitude for shaping and transforming microbial populations (Mougin & Joyce 2022). Notwithstanding, self-assembling virus-like polymers composed of viral protein coat monomers have been used as efficacious vaccines (Thiéry et al. 2006).
Next time we dedicate part VII to the innovative research of Gallet and collaborators (2023) who simulated in vitro blooms of toxigenic cyanobacteria while screening the symbiomes, pathobiomes, and metabolomes of healthy, eutrophication, and bloom-exposed fish.
References
Adams, J. (2018) Interesting Ecology Shift of Blacktail and Orangeface Butterflyfish. ReefBuilders.com. https://reefbuilders.com/2018/12/18/blacktail-and-redface-butterflyfish/
Akhter, N., Wu, B., Memon, A., M. & Mohsin, M. (2015) Probiotics and prebiotics associated with aquaculture: a review. Fish Shellfish Immunol. 45, 733-741. https://www.doi.org/10.1016/j.fsi.2015.05.038
Amann, R., I., Ludwig, W. & Schleifer, K., H. (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143-169.
Andreoni, F. & Magnani, M. (2014) Photobacteriosis: Prevention and Diagnosis. Journal of immunology research. 2014,.
Aslett, C., G. (2024a) Coral Immunity Part I. https://www.reefranch.co.uk/
Aslett, C., G. (2024b) Holosystemics Part IV: Dysbiosis and the Microscopic Coral Alliance. https://www.reefranch.co.uk/
Aslett, C., G. (2023c) Fish Immunostimulation. https://www.reefranch.co.uk/
Aslett, C., G. (2024) The Complete Reef Aquarist, for hobbyists, the trade and academics – A Conservation Manual. Reef Ranch Publishing Ltd, Leeds, West Yorkshire, UK.
Austin, B. & Austin, D., A. (2016) Bacterial fish pathogens, 6th ed. Springer, Stirling, UK
Austin, B. (1982) Taxonomy of bacteria isolated from a coastal, marine fish-rearing unit. J. Appl. Bacteriol. 53, 253-268. https://www.doi.org/10.1111/j.1365-2672.1982.tb04684.x
Bass, D., Stentiford. G., D., Wang, H-C., Koskella, B. & Tyler, C., R. (2019) The pathobiome in animal and plant diseases. Trends Ecol Evol. 34(11), 996-1008.
Bell, G., R., Hoskins, G., E. & Hodgkiss, W. (1971) Aspects of the characterization, identification, and ecology of the bacterial flora associated with the surface of stream-incubating Pacific salmon (Oncorhynchus) eggs. J. Fish. Board Can. 28, 1511-1525. https://www.doi.org/10.1139/f71-232
Birrell, C., Mccook, L., Willis, B. & Diaz-Pulido, G. (2008) Effects of benthic algae on the replenishment of corals and the implications for the resilience of coral reefs. Oceanography and marine biology. 46,.
Blazer, V., Phillips, S. & Pendleton, E. (2016) Fish Health, Fungal Infections, and Pfiesteria: The Role of the U.S. Geological Survey. U.S. Geological Survey Fact Sheet. pp 114-198.
Boilard, A., Dubé, C., E., Gruet, C., Mercière, A., Hernandez-Agreda, A. & Derome, N. (2020) Defining Coral Bleaching as a Microbial Dysbiosis within the Coral Holobiont. Microorganisms. 8, 1682. https://doi.org/10.3390/microorganisms8111682
Bone, Q., Marshall, N., B. & Blaxter, J., H., S. (1995) Biology of Fishes. Glasgow: Blackie Academic & Professional. https://www.doi.org/10.1007/978-1-4615-2664-3
Bozzi, D., Rasmussen, J., A., Carøe, C., et al. (2021) Salmon gut microbiota correlates with disease infection status: potential for monitoring health in farmed animals. Anim Microbiome. 3(1), 1-17.
Brandley, B., K. & Schnaar, R., L. (1986) Cell-Surface Carbohydrates in Cell Recognition and Response. J Leukoc Biol 40, 97–111. https://doi.org/10.1002/jlb.40.1.97
Brown, R., Moore, L., Mani, A., Patel, S., Salinas, I. (2021) Effects of ploidy and salmonid alphavirus infection on the skin and gill microbiome of Atlantic salmon (Salmo salar). PLoS One. 16(2), e0243684.
Cámara-Ruiz, M., Cerezo, I., M., Guardiola, F., A. et al. (2021) Alteration of the immune response and the microbiota of the skin during a natural infection by Vibrio harveyi in European seabass (Dicentrarchus labrax). Microorganisms. 9(5), 964.
Carnevali, O., Maradonna, F. & Gioacchini, G. (2017) Integrated control of fish metabolism, wellbeing and reproduction: the role of probiotic. Aquaculture 472, 144-155. https://www.doi.org/10.1016/j.aquaculture.2016.03.037
Cerezuela, R., Meseguer, J. & Esteban, M. (2011) Current knowledge in synbiotic use for fish aquaculture: a review. J. Aquac. Res. Dev. 1, 1-7.
Cherrak, Y., Flaugnatti, N., Durand, E., Journet, L. & Cascales, E. (2019) Structure and Activity of the Type VI Secretion System. Microbiology spectrum, 7(4), 1.
Colorni, A., Avtalion, R., Knibb, W., Berger, E., J., Colorni, B. & Timan, B. (1998) Histopathology of sea bass (Dicentrarchus labrax) experimentally infected with Mycobacterium marinum and treated with streptomycin and garlic (Allium sativum) extract. Aquaculture. 160, 1-17.
Cordero, H., Guardiola, F., A., Tapia-Paniagua, S., T., Cuesta, A., Meseguer, J., Balebona, M., C., et al. (2015) Modulation of immunity and gut microbiota after dietary administration of alginate encapsulated Shewanella putrefaciens Pdp11 to gilthead seabream (Sparus aurata L.). Fish Shellfish Immunol. 45, 608-618. https://www.doi.org/10.1016/j.fsi.2015.05.010
Das, P., Mandal, S., Khan, A., Manna, S., K. & Ghosh, K. (2014) Distribution of extracellular enzyme-producing bacteria in the digestive tracts of 4 brackish water fish species. Turk. J. Zool. 38, 79-88. https://www.doi.org/10.3906/zoo-1205-3
Delcroix, J., Gatesoupe, F., J., Desbruyères, E., Huelvan, C., Le Delliou, H., Le Gall, M., M., Quazuguel, P., et al. (2015) The effects of dietary marine protein hydrolysates on the development of sea bass larvae, Dicentrarchus labrax, and associated microbiota. Aquac. Nutr. 21, 98-104. https://www.doi.org/10.1111/anu.12139
Duperron, S., Halary, S., Habiballah, M., Gallet, A., Huet, H., Duval, C. et al. (2019) Response of fish gut microbiota to toxin-containing cyanobacterial extracts: a microcosm study on the Medaka (Oryzias latipes). Environ SciTechnol Lett. 6, 341–7.
Egerton, S., Culloty, S., Whooley, J., Stanton, C. & Ross, R., P. (2018) The Gut Microbiota of Marine Fish. Frontiers in Microbiology. 9(873),. https://www.frontiersin.org/articles/https://doi.org/10.3389/fmicb.2018.00873/full
Elhassan, Y., Philp, A. & Lavery, G. (2017) Targeting NAD+ in Metabolic Disease: New Insights Into an Old Molecule. Journal of the Endocrine Society. 1(7), 816-835.
Estruch, G., Collado, M., Peñaranda, D., Vidal, A., T., Cerdá, M., J., Martínez, G., P., et al. (2015) Impact of fishmeal replacement in diets for gilthead sea bream (Sparus aurata) on the gastrointestinal microbiota determined by pyrosequencing the 16S rRNA gene. PLoS One 10, e0136389. https://www.doi.org/10.1371/journal.pone.0136389
Flerova, E. & Balabanova, L. (2013) Ultrastructure of granulocytes of teleost fish (Salmoniformes, Cypriniformes, Perciformes). Journal of Evolutionary Biochemistry and Physiology. 49(2), 223-233.
Fournier, V., Huelvan, C. & Desbruyeres, E. (2004) Incorporation of a mixture of plant feedstuffs as substitute for fish meal in diets of juvenile turbot (Psetta maxima). Aquaculture 236, 451-465. https://www.doi.org/10.1016/j.aquaculture.2004. 01.035
Gallet, A., Halary, S., Duval, C., Huet, H., Duperron, S. & Marie, B. (2023) Disruption of fish gut microbiota composition and holobiont’s metabolome during a simulated Microcystis aeruginosa (Cyanobacteria) bloom. Microbiome. 11, 108. https://doi.org/10.1186/s40168-023-01558-2
Ghanbari, M., Kneifel, W. & Domig, K., J. (2015) A new view of the fish gut microbiome: advances from next-generation sequencing. Aquaculture 448, 464-475. https://www.doi.org/10.1016/j.aquaculture.2015.06.033
Giatsis, C., Sipkema, D., Smidt, H. et al. (2015) The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci Rep. 5(1), 1-15.
Green, T., J., Smullen, R. & Barnes, A., C. (2013) Dietary soybean protein concentrate-induced intestinal disorder in marine farmed Atlantic salmon, Salmo salar is associated with alterations in gut microbiota. Vet. Microbiol. 166, 286-292. https://www.doi.org/10.1016/j.vetmic.2013.05.009
Hansen, G. & Olafsen, J. (1999) Bacterial interactions in early life stages of marine cold water fish. Microb. Ecol. 38, 1-26. https://www.doi.org/10.1007/s002489900158
Hlongwane, P., Mungra, N., Madheswaran, S., Akinrinmade, O., A., Chetty, S. & Barth, S. (2018) Human Granzyme B Based Targeted Cytolytic Fusion Proteins. Biomedicines. 6(2), 72.
Ho, J. & Kim, I. (2001) New species of Hatschekia Poche, 1902 (Copepoda: Hatschekiidae) parasitic on marine fishes of Kuwait. Syst Parasitol. 49, 73-79.
Hooper, L., V. & Gordon, J., I. (2001) Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11, 1R-10R. https://doi.org/10.1093/glycob/11.2.1R
Hovanec, T. (2019) Dr. How to harness bacteria to cycle your saltwater tank quickly! | MACNA 2019. BrsTV. https://www.youtube.com/watch?v=zDI7sxqC-ss
James, A., G. (1988) Are clupeid microphagists herbivorous or omnivorous? A review of the diets of some commercially important clupeids. S. Afr. J. Mar. Sci. 7, 161-177. https://www.doi.org/10.2989/025776188784379017
Kapoor, B. & Khawna, B. (1993) The potential spectrum of the gut in teleost fishes. Adv. Fish Res. 1, 221-226.
Karachle, P., K. & Stergiou, K., I. (2010) Gut length for several marine fish: relationships with body length and trophic implications. Mar. Biodivers. Rec. 3, e106. https:///www.doi.org/10.1017/S1755267210000904
Kelly, C. & Salinas, I. (2017) Under pressure: Interactions between commensal microbiota and the teleost immune system. Front. Immunol. 8, 1.
Kim, B.,-R., Shin. J., Guevarra, R., B., et al. (2017) Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol. 27(12), 2089-2093.
Kim, J. & Lee, J., L. (2017) Correlation of Total Bacterial and Vibrio spp. Populations between Fish and Water in the Aquaculture System. Frontiers in Marine Science. 4,.
Kiron, V. (2012) Fish immune system and its nutritional modulation for preventive health care. Anim. Feed Sci. Technol. 173, 111-133. https://www.doi.org/10.1016/j.anifeedsci.2011.12.015
Kline, D., I., Kuntz, N., M., Breitbart, M., Knowlton, N. & Rohwer, F. (2006) Role of elevated organic carbon levels and microbial activity in coral mortality. Marine Ecology Progress Series. 314, 119-125.
Langille, M., G., Zaneveld, J., Caporaso, J., G. et al. (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 31(9), 814-821.
Larsen, A., M. (2014) Studies on the Microbiota of Fishes and the Factors Influencing Their Composition. Auburn, AL: Auburn University.
Lasica, A., Ksiazek, M., Madej, M. & Potempa, J. (2017) The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function. Frontiers in Cellular and Infection Microbiology. 7,.
Legrand, T., P., R., A., Catalano, S., R., Wos-Oxley, M., L., Stephens, F., Landos, M., Bansemer, M., S., Stone, D., A., J., Qin, J., G. & Oxley, A., P., A. (2018) The Inner Workings of the Outer Surface: Skin and Gill Microbiota as Indicators of Changing Gut Health in Yellowtail Kingfish. Front. Microbiol. 8:2664. https://www.doi.org/10.3389/fmicb.2017.02664
Lesel, R., De La Noüe, J. & Choubert, G. (1989) Fecal bacterial flora of rainbow trout under antibiotic treatment: effect of the number of pyloric caeca and the lipid content of food. Aquaculture: A Biotechnology in Progress, Vol. 1. De Pauw, N., Jaspers, E., Ackefors, H. & Wilkins, N. (eds.). Bredene: European Aquaculture Society, 592.
Li, P. & Gatlin, D., M. (2003) Evaluation of brewers yeast (Saccharomyces cerevisiae) as a feed supplement for hybrid striped bass (Morone chrysopsX—M. saxatilis). Aquaculture. 219(1), 681-692.
Li, T., Li, H., Gatesoupe, F-J. et al. (2017) Bacterial signatures of ‘red-operculum’ disease in the gut of crucian carp (Carassius auratus). Microb Ecol. 74(3), 510-521.
Liu, Q., Lai, Z., Gao, Y. et al. (2021) Connection between the gut microbiota of largemouth bass (Micropterus salmoides) and microbiota of the pond culture environment. Microorganisms. 9(8), 1770.
Llewellyn, M., Leadbeater, S., Garcia, C. et al. (2017) Parasitism perturbs the mucosal microbiome of Atlantic Salmon. Sci Rep. 7(1), 1-10.
Llewellyn, M., S., Boutin, S., Hoseinifar, S., H. & Derome, N. (2018) Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front Microbiol. 2014, 5. http://journal.frontiersin.org/article/10.3389/fmicb.2014.00207/abstract
Lobo, C., Moreno-Ventas, X., Tapia-Paniagua, S., Rodríguez, C., Moriñigo, M., A. & de La Banda, I., G. (2014) Dietary probiotic supplementation (Shewanella putrefaciens Pdp11) modulates gut microbiota and promotes growth and condition in Senegalese sole larviculture. Fish Physiol. Biochem. 40, 295-309. https://www.doi.org/10.1007/s10695-013-9844-0
Lom, J. & Corliss, J. (1971) Morphogenesis and Cortical Ultrastructure of Brooklynella hostilis, a Dysteriid Ciliate Ectoparasitic on Marine Fishes. The Journal of Eukaryotic Microbiology. 18(2), 261-281.
López Nadal, A., Peggs, D., Wiegertjes, G., F. & Brugman, S. (2018) Exposure to Antibiotics Affects Saponin ImmersionInduced Immune Stimulation and Shift in Microbial Composition in Zebrash Larvae. Front Microbiol 9, 2588. https://doi.org/10.3389/fmicb.2018.02588
López-Dóriga, M., Barnes, A., dos Santos, N. & Ellis, A. (2000) Invasion of fish epithelial cells by Photobacterium damselae subsp. piscicida: evidence for receptor specificity, and effect of capsule and serum. Microbiology. 146(1), 21-30.
Lorgen-Ritchie, M., Clarkson, M., Chalmers, L., Taylor, J., F., Migaud, H. & Martin, S., A. M. (2021) A temporally dynamic gut microbiome in Atlantic salmon during freshwater recirculating aquaculture system (RAS) production and post-seawater transfer. Front. Mar. Sci. 8, 869. https://doi.org/10.3389/fmars.2021.711797
Lorgen-Ritchie, M., Webster, T., U., McMurtrie, J., Bass, D., Tyler, C., R., Rowley, A. & Martin, S., A., M. (2023) Microbiomes in the context of developing sustainable intensified aquaculture. Frontiers in Microbiology. 14, 1200997.
Ma, C., Chen, C., Jia, L., He, X. & Zhang, B. (2019) Comparison of the intestinal microbiota composition and function in healthy and diseased Yunlong Grouper. AMB Express. 9(1), 1-11.
Martins, J. & Vasconcelos, V. (2015) Cyanobactins from cyanobacteria: current genetic and chemical state of knowledge. Mar Drugs. 13, 6910-6946.
Mashoof, S. & Criscitiello, M., F. (2016) Fish Immunoglobulins. Biology. 5(4), 45.
McBride, M., J. & Nakane, D. (2015) Flavobacterium gliding motility and the type IX secretion system. Curr. Opin. Microbiol. 28, 72-77.
McCauley, M., Goulet, T., L., Jackson, C., R. & Loesgen, S. (2022) Systematic review of cnidarian microbiomes reveals insights into the structure, specificity, and fidelity of marine associations. Nature Communications. 14, 4899. https://doi.org/10.1038/s41467-023-39876-6
Merrifield, D., L. & Rodiles, A. (2015) The fish microbiome and its interactions with mucosal tissues. Mucosal Health in Aquaculture. Peatman, E. (ed.) Academic Press, San Diego, CA. pp 273-295.
Mihalitsis, M. & Bellwood, D. (2017) A morphological and functional basis for maximum prey size in piscivorous fishes. PLoS ONE. 12(9),.
Miyake, S., Ngugi, D. K. & Stingl, U. (2015) Diet strongly influences the gut microbiota of surgeonfishes. Mol. Ecol. 24, 656-672. https://www.doi.org/10.1111/mec.13050
Miyake, S., Soh, M., Azman, M., N., Ngoh, S., Y., Orbán, L., Seedorf, H. (2020) Insights into the microbiome of farmed Asian sea bass (Lates cal-carifer) with symptoms of tenacibaculosis and description of Tenacibaculum singaporense sp. nov. Antonie Van Leeuwenhoek. 113(6), 737-752.
Monroig, Ó., Tocher, D., R. & Navarro, J., C. (2013) Biosynthesis of polyunsaturated fatty acids in marine invertebrates: recent advances in molecular mechanisms. Mar. Drugs 11, 3998-4018. https://www.doi.org/10.3390/md11103998
Moon, D. (2021) Boosting NAD+ to Reverse Aging? Overview of NR and NMN. GeneticLifeHacks.com. https://www.geneticlifehacks.com/nad-reversing-aging-overview-of-nr-and-nmn/
Mouchet, M., A., Bouvier, C., Bouvier, T., Troussellier, M., Escalas, A. & Mouillot, D. (2012) Genetic difference but functional similarity among fish gut bacterial communities through molecular and biochemical fingerprints. FEMS Microbiol. Ecol. 79, 568-580. https://www.doi.org/10.1111/j.1574-6941.2011. 01241.x
Mougin, J. & Joyce, A. (2022) Reviews in Aquaculture. Fish disease prevention via microbial dysbiosis-associated biomarkers in aquaculture. 15, 579-594. https://doi.org/10.1111/raq.12745
Muñoz-Baquero, M., Lorenzo-Rebenaque, L., Garc´ıa-Va´zquez, F., A., Garc´ıa-Pa´rraga, D., Mart´ınez-Priego, L., De Marco-Romero, G., Gala´n-Vendrell, I., D’Auria, G. & Marco-Jime´nez, F. (2023) Unveiling Microbiome Signature In Inner Body Fluids: Comparison Between Wild And Aquarium Small-Spotted Catshark (Scyliorhinus canicular). Frontiers in Marine Science. https://www.doi.org/10.3389/fmars.2023.1151119
Murdoch, C., C. & Rawls, J., F. (2019) Commensal microbiota regulate vertebrate innate immunity-insights from the zebrafish. Front. Immunol. 10:2100. https://doi.org/10.3389/fimmu.2019.02100
Navarrete, P., Espejo, R. T. & Romero, J. (2009) Molecular analysis of microbiota along the digestive tract of juvenile Atlantic salmon (Salmo salar L.). Microb. Ecol. 57, 550-561. https://www.doi.org/10.1007/s00248-008-9448-x
Naya-Català, F., Piazzon, M., C., Calduch-Giner, J. A., Sitjà-Bobadilla, A. & Pérez-Sánchez, J. (2022) Diet and host genetics drive the bacterial and fungal intestinal metatranscriptome of Gilthead Sea bream. Front. Microbiol. 13, 883738. https://doi.org/10.3389/fmicb.2022.883738
Nayak, S., K. (2010) Role of gastrointestinal microbiota in fish. Aquac. Res. 41, 1553-1573. https://www.doi.org/10.1111/j.1365-2109.2010.02546.x
Neuman, C., Hatje, E., Zarkasi, K. Z., Smullen, R., Bowman, J., P. & Katouli, M. (2016) The effect of diet and environmental temperature on the faecal microbiota of farmed Tasmanian Atlantic Salmon (Salmo salar L.). Aquac. Res. 47, 660-672. https://www.doi.org/10.1111/are.12522
Noga, E., J. (2010) Fish Disease. Diagnosis and Treatment. Second Edition. Wiley-Blackwell, John Wiley & Sons Inc. p 139.
Oliva-Teles, A. & Goncalves, P. (2001) Partial replacement of fishmeal by brewers yeast (Saccaromyces cerevisae) in diets for sea bass (Dicentrarchus labrax) juveniles. Aquaculture. 202(3), 269-278.
Orlandi, I., Alberghina, L. & Vai, M. (2020) Nicotinamide, Nicotinamide Riboside and Nicotinic Acid-Emerging Roles in Replicative and Chronological Aging in Yeast. Biomolecules. 10(4), 604.
Ortiz-Estrada, A., M., Gollas-Galván, T., Martínez-Cordova, L., R. & Martínez-Porchas, M. (2019) Predictive functional profiles using metagenomic 16S rRNA data: a novel approach to understanding the microbial ecology of aquaculture systems. Rev Aquacult. 11(1), 234-245.
Perdiguero, P., Martin-Martin, A., Benedicenti, O., Diaz-Rosales, P., Morel, E., Munoz-Atienza, E., Garcia-Flores, M., Simon, R., Soleto, I., Cerutti, A. & Tafalla, C. (2019) Teleost IgD+Ig- B Cells Mount Clonally Expanded and Mildly Mutated Intestinal IgD Responses in the Absence of Lymphoid Follicles. Cell Reports. 29(13), 4223-4235.
Pérez-Pascual, D., Lunazzi, A., Magdelenat, G., Rouy, Z., Roulet, A., Lopez-Roques, C., Larocque, R., Barbeyron, T., Gobet, A., Michel, G., Bernardet, J. & Duchaud, E. (2017) The Complete Genome Sequence of the Fish Pathogen Tenacibaculum maritimum Provides Insights into Virulence Mechanisms. Frontiers in Microbiology. 8,.
Pollock, F.,J., McMinds, R., Smith, S., Bourne, D., G., Willis, B., L., Medina, M., Thurber, R., V. & Zaneveld, J., R. (2018) Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun. 9, 4921.
Reid, K., M., Patel, S., Robinson, A., J., et al. (2017) Salmonid alphavirus infection causes skin dysbiosis in Atlantic salmon (Salmo salar L.) post-smolts. PLoS One. 12(3):e0172856.
Richardson, L., L., Sekar, R., Myers, J., L., Gantar, M., Voss, J., D., Kaczmarsky, L., Remily, E., R., Boyer, G., L. & Zimba, P., V. (2007) The presence of the cyanobacterial toxin microcystin in black band disease of corals. FEMS Microbiology Letters. 272(2), 182-187.
Riley, M., A. & Wertz, J., E. (2002) Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 84, 357-364.
Ringø, E., Lødemel, J., B., Myklebust, R., Jensen, L., Lund, V., Mayhew, T., M., et al. (2002) The effects of soybean, linseed and marine oils on aerobic gut microbiota of Arctic charr Salvelinus alpinus L. before and after challenge with Aeromonas salmonicida ssp. salmonicida. Aquac. Res. 33, 591-606. https://www.doi.org/10.1046/j.1365-2109.2002.00697.x
Ringø, E., Zhou, Z., Vecino, J., L., G., Wadsworth, S., Romero, J., Krogdahl, Å., et al. (2016) Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac. Nutr. 22, 219-282. https://www.doi.oeg/10.1111/anu.12346
Ritchie, K., B. & Smith, G., W. (1995) Preferential carbon utilization by surface bacterial communities from water mass, normal, and white-band diseased Acropora cervicornis. Mol Mar Biol Biotechnol. 4, 345-354.
Rohde, K. (1932) Ciliophora (ciliates). Marine Parasitology. Rohde, K. (ed.). CSIRO Publishing, Clayton, Australia.
Rombout, J., H., Abelli, L., Picchietti, S., Scapigliati, G. & Kiron, V. (2011) Teleost intestinal immunology. Fish Shellfish Immunol. 31, 616-626. https://www.doi.org/10.1016/j.fsi. 2010.09.001
Rosado, D., Pérez-Losada, M., Pereira, A., Severino, R. & Xavier, R. (2021) Effects of aging on the skin and gill microbiota of farmed seabass and seabream. Anim Microbiome. 3(1), 1-14.
Rosado, D., Pérez-Losada, M., Severino, R. & Xavier, R. (2022) Monitoring infection and antibiotic treatment in the skin microbiota of farmed European seabass (Dicentrarchus Labrax) fingerlings. Microb Ecol. 83(3), 789-797.
Rosado, D., Perez-Losada, M., Severino, R., Cable, J. & Xavier, R. (2019b) Characterization of the skin and gill microbiomes of the farmed seabass (Dicentrarchus labrax) and seabream (Sparus aurata). Aquaculture. 500, 57-64.
Rosado, D., Xavier, R., Severino, R., Tavares, F., Cable, J. & Pérez-Losada, M. (2019a) Effects of disease, antibiotic treatment and recovery trajectory on the microbiome of farmed seabass (Dicentrarchus labrax). Sci Rep. 9(1), 1-11.
Rozas-Serri, M. (2019) Gill diseases in marine salmon aquaculture with an emphasis on amoebic gill disease. CAB Reviews Perspectives in Agriculture Veterinary Science Nutrition and Natural Resources. 14, 1-15.
Santoro, E., P., Borges, R., M., Espinoza, J., L., Freire, M., Messias, C., S., M., A., Villela, H., D., M., Pereira, L., M., Vilela, C., L., S., Rosado, J., G., Cardoso, P., M., Rosado, P., M., Assis, J., M., Duarte, G., A., S., Perna, G., Rosado, A., S., Macrae, A., Dupont, C., L., Nelson, K., E., Sweet, M., J., Voolstra, C., R. & Peixoto, R., S. (2021) Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Science advances. 7(33), 3088.
Santos, F., F., Yamamoto, D., Abe, C., M., Bryant, J., A., Hernandes, R., T., Kitamura, F., C., Castro, F., S., Valiatti, T., B., Piazza, R., Elias, W., P., Henderson, I., R. & Gomes, T. (2019) The Type III Secretion System (T3SS)-Translocon of Atypical Enteropathogenic Escherichia coli (aEPEC) Can Mediate Adherence. Frontiers in microbiology. 10, 1527.
Scapigliati, G., Fausto, A., M. & Picchietti, S. (2018) Fish Lymphocytes: An Evolutionary Equivalent of Mammalian Innate-Like Lymphocytes? Frontiers in immunology. 9, 971.
Segata, N., Izard, J., Waldron, L., et al. (2011) Metagenomic biomarker discovery and explanation. Genome Biol. 12(6), 1-18.
Shabir, U., Ali, S., Magray, A., Ganai, B., Firdous, P., Hassan, T. & Nazir, R. (2018) Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microbial pathogenesis. 114, 50-56.
Sila, A., Nedjar-Arroume, N., Hedhili, K., Chataigné, G., Balti, R., Nasri, M., et al. (2014) Antibacterial peptides from barbel muscle protein hydrolysates: activity against some pathogenic bacteria. LWT Food Sci. Technol. 55, 183-188. https://www.doi.org/10.1016/j.lwt.2013.07.021
Siriyappagouder, P., Galindo-Villegas, J., Dhanasiri, A., K., S., Zhang, Q., Mulero, V., Kiron, V. et al. (2020) Pseudozyma priming influences expression of genes involved in metabolic pathways and immunity in zebrafish larvae. Front. Immunol. 11, 987. https://doi.org/10.3389/fimmu.200.00978
Siriyappagouder, P., Kiron, V., Lokesh, J., Rajeish, M., Kopp, M. & Fernandes, J. (2018) The intestinal mycobiota in wild zebrafish comprises mainly Dothideomycetes while Saccharomycetes predominate in their laboratory-reared counterparts. Front. Microbiol. 9, 387. https://doi.org/10.3389/fmicb.2018.00387
Sitjà-Bobadilla, A., Gil-Solsona, R., Estensoro, I., Piazzon, M., Martos-Sitcha, J., Picard-Sánchez, A., Fuentes, J., Sancho, J., Calduch-Giner, J., Hernández, F. & Pérez-Sánchez, J. (2019) Disruption of gut integrity and permeability contributes to enteritis in a fish-parasite model: a story told from serum metabolomics. Parasites & Vectors. 12,.
Skrodenyte-ArbaCIauskiene, V. (2007) Enzymatic activity of intestinal bacteria in roach Rutilus rutilus L. Fish. Sci. 73, 964-966. https://www.doi.org/10.1111/j.1444-2906.2007. 01421.x
Slinger, J., Adams, M., B., Stratford, C., N., Rigby, M. & Wynne, J., W. (2021) The effect of antimicrobial treatment upon the gill bacteriome of Atlantic salmon (Salmo salar L.) and progression of amoebic gill disease (AGD) in vivo. Microorganisms. 9, 987. https://doi.org/10.3390/microorganisms9050987
Spanova, M. & Daum, G. (2011) Squalene–biochemistry, molecular biology, process biotechnology, and applications. Eur J Lipid Sci Technol. 113(11), 1299-1320.
Spor, A., Koren, O. & Ley, R. (2011) Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279-290. https://doi.org/10.1038/nrmicro2540
Stevens, C., E. & Hume, I., D. (2004) Comparative Physiology of the Vertebrate Digestive System. Cambridge: Cambridge University Press.
Stevens, J., Jackson, R., Olson, J. (2016) Bacteria associated with lionsh (Pterois volitans/miles complex) exhibit antibacterial activity against known fish pathogens. Mar Ecol Prog Ser 558, 167–180. https://doi.org/10.3354/meps11789
Sullam, K., E., Essinger, S., D., Lozupone, C., A., O’Connor, M., P., Rosen, G., L., Knight, R., et al. (2012) Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21, 3363-3378. https://www.doi.org/10.1111/j.1365-294X.2012.05552.x
Swiatecka, D., Markiewicz, L., H. & Wróblewska, B. (2012) Pea protein hydrolysate as a factor modulating the adhesion of bacteria to enterocytes, epithelial proliferation and cytokine secretion–an in vitro study. Cent. Eur. J. Immunol. 37, 227-231. https://www.doi.org/10.5114/ceji.2012.3079
Tapia-Paniagua, S., T., Ceballos-Francisco, D., Balebona, M., C., Esteban, M., A. & Moriñigo, M., A. (2018) Mucus glycosylation, immunity and bacterial microbiota associated to the skin of experimentally ulcered gilthead seabream (Sparus aurata). Fish Shellfish Immunol. 75, 381-390.
Thiéry, R., Cozien, J., Cabon, J., Lamour, F., Baud, M. & Schneemann, A. (2006) Induction of a Protective Immune Response against Viral Nervous Necrosis in the European Sea Bass Dicentrarchus labrax by Using Betanodavirus Virus-Like Particles. Journal of Virology. 80(20), 10201.
Torrecillas, S., Montero, D. & Izquierdo, M. (2014) Improved health and growth of fish fed mannan oligosaccharides: potential mode of action. Fish Shellfish Immunol. 36, 525-544. https://doi.org/10.1016/j.fsi.2013.12.029
Tretina, K., Park, E., S., Maminska, A. & MacMicking, J., D. (2019) Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. The Journal of Experimental Medicine. 216(3), 482-500.
Valero, Y., Saraiva‐Fraga, M., Costas, B. & Guardiola, F., A. (2019) Antimicrobial peptides from fish: beyond the fight against pathogens. Reviews in Aquaculture. 12, 224-253.
Vayssier-Taussat, M., Albina, E., Citti, C., et al. (2014) Shifting the paradigm from pathogens to pathobiome: new concepts in the light of metaomics. Front Cell Infect Microbiol. 4, 29.
Vestrum, R., I., Forberg, T., Luef, B., Bakke, I., Winge, P., Olsen, Y., et al. (2022) Commensal and opportunistic bacteria present in the microbiota in Atlantic cod (Gadus morhua) larvae differentially alter the hosts’ innate immune responses. Microorganisms. 10, 24. https://doi.org/10.3390/microorganisms10010024
Walke, J., B., et al. (2017) Dominance-function relationships in the amphibian skin microbiome. Environ. Microbiol. 19, 3387–3397.
Wang, S. & Loreau, M. (2014) Ecosystem stability in space: α, β and γ variability. Ecol Lett. 17(8), 891-901.
Webster, T., M., U., Rodriguez-Barreto, D., Consuegra, S. & Garcia de Leaniz, C. (2019) Cortisol-induced signatures of stress in the fish microbiome. bioRxiv:826503. https://doi.org/10.1101/826503
Wilson, J., M. & Castro, L., F., C. (2010) Morphological diversity of the gastrointestinal tract in fishes. Fish Physiol. 30, 1-55. https://www.doi.org/10.1016/S1546-5098(10)03001-3
Wilson, M. World Feeds Limited, 3b Coulman Street Industrial Estate, Thorne, Doncaster, DN8 5JS, United Kingdom.
Wu, J., Mao, C., Deng, Y., et al. (2019) Diversity and abundance of antibiotic resistance of bacteria during the seedling period in marine fish cage-culture areas of Hainan, China. Mar Pollut Bull 141:343–349. https://doi.org/10.1016/j.marpolbul.2019.02.069
Xavier, R., Pereira, A., Pagan, A., Hendrick, G., C., Nicholson, M., D., Rosado, D., Soares, M., C., Pérez-Losada, M., Sikkel, P., C. (2020) The effects of environment and ontogeny on the skin microbiome of two Stegastes damselfishes (Pomacentridae) from the eastern Caribbean Sea. Mar Biol. 167(7),1-12.
Xu, Z., Takizawa, F., Casadei, E., Shibasaki, Y., Ding, Y., Sauters, T., J., C., et al. (2020) Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol. 5, 3254. https://doi.org/10.1126/sciimmunol.aay3254
Yatsunenko, T., Rey, F., E., Manary, M., J., Trehan, I., Dominguez-Bello, M., G., Contreras, M., et al. (2012) Human gut microbiome viewed across age and geography. Nature 486, 222-227. https://www.doi.org/10.1038/nature11053
Yoon, J., Matsuo, Y., Matsuda, S., Adachi, K., Kasai, H. & Yokota, A. (2018) Rubritalea sabuli sp. nov., a carotenoid-and squalene-producing member of the family Verrucomicrobiaceae, isolated from marine sediment. Int J Syst Evol Microbiol. 58(4), 992-997.
Yoshimizu, M., Kimura, T. & Sakai, M. (1980) Microflora of the embryo and the fry of salmonids. Bull. Jpn. Soc. Sci. Fish. 46, 967-975. https://www.doi.org/10.2331/suisan.46.967
Yu, Y., Wang, Q., Huang, Z., Ding, L. & Xu, Z. (2020) Immunoglobulins, Mucosal Immunity and Vaccination in Teleost Fish. Frontiers in immunology. 11, 567941.
Zanefeld, J., R. et al. (2017) Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2, 17121.
Zarkasi, K., Z., Taylor, R., S., Abell, G., C., Tamplin, M., L., Glencross, B., D. & Bowman, J., P. (2016) Atlantic salmon (Salmo salar L.) gastrointestinal microbial community dynamics in relation to digesta properties and diet. Microb. Ecol. 71, 589-603. https://www.doi.org/10.1007/s00248-015-0728-y
Zeng, A., Tan, K., Gong, P. et al. (2020) Correlation of microbiota in the gut of fish species and water. 3 Biotech. 0(11), 1-10.
Zhang, L., Ni, C., Xu, W., Dai, T., Yang, D., Wang, Q., Zhang, Y. & Liu, Q. (2016) Intramacrophage Infection Reinforces the Virulence of Edwardsiella tarda. Journal of bacteriology. 198(10), 1534-1542.
Zhang, M., Shan, C., Tan, F., Limbu, S., M., Chen, L. & Du, Z. (2020) Gnotobiotic models: powerful tools for deeply understanding intestinal microbiota-host interactions in aquaculture. Aquaculture 517:734800. https://doi.org/10.1016/j.aquaculture.2019.734800
Zhang, X., Ding, L., Yu, Y. et al. (2018) The change of teleost skin commensal microbiota is associated with skin mucosal transcriptomic responses during parasitic infection by Ichthyophthirius multifillis. Front Immunol. 9, 2972.
Zhang, Y. & Gui, J. (2012) Molecular regulation of interferon antiviral response in fish. Developmental and comparative immunology. 38(2), 193-202.
Zhou, Z., Liu, Y., Shi, P., He, S., Yao, B. & Ringø, E. (2009) Molecular characterization of the autochthonous microbiota in the gastrointestinal tract of adult yellow grouper (Epinephelus awoara) cultured in cages. Aquaculture 286, 184-189. https://www.doi.org/10.1016/j.aquaculture.2008.10.002

Hashtags: #coralreefs #coralconservation #reefrestoration #MarineConservation #coralreefconservation #oceanconservation #thecompletereefaquarist #reefconservation #reefaquarium; Metaphrases: coral reef expert*, marine aquarium guide, reef tank fish guide, reef tank guide, saltwater aquarium guide

0 Comments