Whereas it may seem investigations of freshwater fish are of little use to reef enthusiasts due to oceanic and riverine disparities, the way salt- and fresh-water microbial consortia and their hosts respond to abiotic and biotic variations are remarkably similar. Furthermore laboratory investigations housing fish in aquaria reliably yield analogous results which repudiates the myth and age-old paradigm that “all systems run differently”.
Blooms of Cyanobacteria including Microcystis aeruginosa cause eutrophication-derived ecological hypoxia in freshwater; and manufacture antimicrobials, enzyme inhibitors, and cytotoxins, while fish are routinely exposed throughout their lifetime. The hepatotoxin microcystin-LR (MC-LR) amasses in liver where it instigates inflammation and necrosis through oxidative stress. Fish are affected after ingestion of contaminated prey or water (Gallet et al. 2023), albeit freshwater fish do not have to drink like saltwater teleosts. Finfish gut microbiota appear less diverse compared to higher vertebrates where the phyla Bacteroidetes, Firmicutes and Proteobacteria predominate (Egerton et al. 2018; Llewellyn et al. 2018). Extracts of various Microcystis strains caused perturbations of fish gut microflora, whereas MC-LR did not (Duperron et al. 2019). “Algae” manufacturer allelopathic compounds that affect the life cycle and the reproductive competence (fecundity) of other species (Birrell et al. 2008; Gallet et al. 2023) whereas microcystin has been isolated from the cyanobacterial mat of Floridian and Bahamian black band disease of corals (Richardson et al. 2007). Cyano-metabolites may exert a more profound effect on microbiomes and the development of a pathobiome (Gallet et al. 2023).
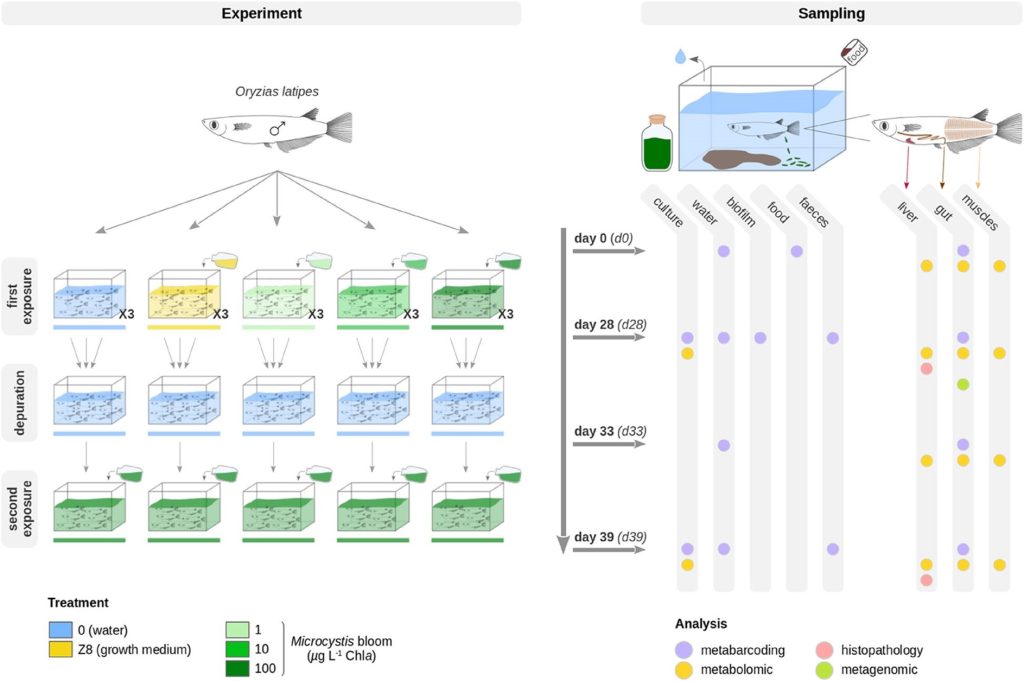
Fig 1. Experiments evaluating the effects of a cyanobacterial bloom of Microcystis aeruginosa on the gut microbial populations of freshwater medaka fish (Oryzias latipes). The first phase lasted 28 days where fish were kept in the growth medium (d28_Z8; yellow), water (d28_0; blue) or the Microcystis aeruginosa chlorophyll a (Chla) densities of 1, 10, and 100 micrograms per litre (µg l-1; ppb) designated d28_1 (light green), d28_10 (medium green), and d28_100 (dark green) which equate to negligeable, ~0.4, and ~10.4 µg l-1 microcystin-LR (MC-LR). All treatments were purged on day 28 in freshwater until day 33, whereafter all fish were exposed to 100 micrograms per litre (µg l-1) of Chla until day 39. Image courtesy of Gallet et al. 2023 and the Creative Commons Licence 4.0. http://creativecommons.org/licenses/by/4.0/
The first phase of freshwater experiments lasted 28 days, where controls included medaka fish (Oryzias latipes) kept in freshwater (d28_0) or the nutrient-enriching (eutrophication-analogous) cyanobacterial growth medium (d28_Z8), whilst the treatments exposed fish to Microcystis aeruginosa chlorophyll a (Chla) densities of 1, 10, and 100 micrograms per litre (µg l-1; d28_1; d28_10; d28_100) which equated to ~0.4 and ~10.4 µg l-1 (ppb) MC-LR in the latter two, whereas the toxin was below detection levels in the former (Fig 1.; Gallet et al. 2023).
The gut mucosal communities of all fish comprised 42 to 219 amplicon sequence variants (ASVs; @microbial lineages), where fish exposed to the growth medium had higher than average richness of ~136 ± 56 which was curtailed in fish kept in just water at ~82 ± 25 ASVs. Fish gut microbes increased marginally in a stepwise fashion in proportion to increased Chla, MC-LR, and Microcystis aeruginosa.
The treatments (d28_1, d28_10, d28_100) and growth medium (d28_Z8) affected microbial community composition and density in comparison to freshwater controls (d28_0), where d28_Z8 had an analogous effect to d28_1 and d28_10 (Gallet et al. 2023). The gut populations in the d28_0 control fish were dominated by 50, 28, 10, and 5 percent of reads belonging to the bacterial phyla Fusobacteriota, Firmicutes, Proteobacteria, and Bacteroidota respectively. The relative abundances of these phyla changed according to treatments however the greatest difference was observed in d28_100 where Firmicutes were found in merely 3 percent of reads. Aeromonas and Flavobacterium species were more abundant in the fish guts of the d28_Z8 and d28_100 groups, while Reyranella species were merely more plentiful in the latter. The Firmicutes genus ZOR0006 was significantly reduced in the fish gut mucosae exposed to 100 µg l-1 Chla, yet this genus was the most plentiful of this phylum in other fish intestinal communities. Cetobacterium species remained core affiliates of all fish ranging from 27 to 48 percent of reads (Gallet et al. 2023).
Negligeable microeukaryotes and Archaea were detected in the gut populations, while the genera Flavobacterium, Aeromonas, Gemmobacter, and Rhizobiales had proliferated in the d28_100 group. The profusions of three Firmicutes assigned genera were decreased in d28_100 compared with d28_0 where ZOR0006 had accounted for nearly 60 precent (Fig 2.; Gallet et al. 2023).
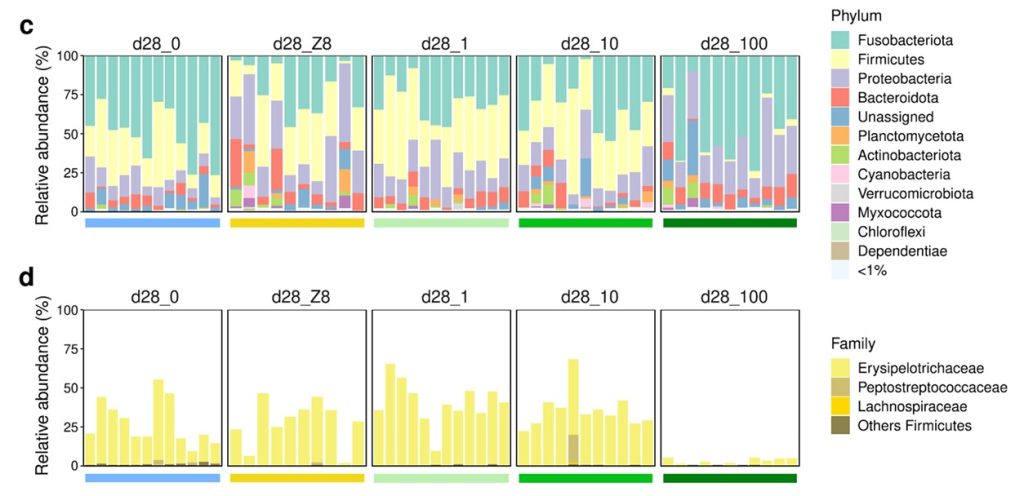
Fig 2. The relative abundances of the bacterial phyla and families of the fish gut communities after the initial 28-day phase of experiments, where d28_0, d28_Z8, d28_1, d28_10, and d28_100 refer to fish kept in freshwater, the growth medium, or the Microcystis aeruginosa Chla densities of 1, 10, and 100 µg l-1 respectively. Image courtesy of Gallet et al. 2023 and the Creative Commons Licence 4.0. http://creativecommons.org/licenses/by/4.0/
The metabolism of a microbiome is referred to as the metabolome, where 1,674 metabolites were identified on day 28. 53 percent of the extracellular products found in medaka fish gut were implicated in the biosynthesis of cobalamin (vitamin B12) like those from Cetobacterium species, whereas ZOR0006 (Erysipelotrichaceae) exhibited carbohydrate-degradative competencies, while both genera could interconvert lactate and pyruvate. Lactate has biocidal properties and is thought vital for gut repair and function despite pyruvate being implicated in the proliferation of pathogens. Gene homologues associated with amino-acid, carbohydrate, and lipid metabolism are common to most prokaryotes, and their products were enriched in the d28_0 and d28_100 groups, whereas enzymes for cofactor and secondary metabolite biosynthesis, and porphyrin and chlorophyll metabolism were amplified in merely the latter. Significant differences were observed between the metabolomes of d28_0 and d28_100 and all other classes, yet those of d28_Z8, d28_1 and d28_10 were comparable (Gallet et al. 2023). Liver metabolites were unaffected yet accumulations in muscle were evident where those from d28_0 and both d28_Z8 and d28_1 were markedly different, whilst those of d28_100 were dissimilar to all other groups except for d28_0 (Gallet et al. 2023).
The least abundant microbes in the d28_0 control group were negatively correlated to several metabolites, where Barnesiellaceae and Epulopiscium were the most plentiful genera, while comprehensive metabolome databases were lacking (Gallet et al. 2023).
Fish were transferred to freshwater on the 28th day as a purgative until day 33 (d33_0_0; d33_Z8_0; d33_1_0; d33_10_0; d33_100_0) whereafter all groups were exposed to 100 µg l-1 Chla Microcystis aeruginosa until day 39 (d39_0_0_100; d39_Z8_0_100; d39_1_0_100; d39_10_0_100; d39_100_0_100). The richness of gut microbiota in these fish decreased from day 28 (82 ± 25) to 33 (50 ± 25), whereafter richness increased until day 39 (72 ± 22). By day 33 all fish gut communities had similar profiles to d28_0, whereas the prokaryotic gut affiliates in d28_100 were mostly identical to d39_100_0_100 (Fig 3.; Gallet et al. 2023).
Microcystis holobiont-associated microbes from water and intestines ranged from 9.8 to 18.1 percent of reads and approached 4.6 percent of reads on day 39 respectively. The communities were somewhat similar between day 33 and day 39 in comparison to the disparity between day 33 and all other treatments, and the difference between day 39 and d28_Z8.
Gut-bound microbes of the phyla Bacteroidota, Fusobacteriota, and Proteobacteria were comparable between day 33 and day 39, whereas Firmicutes were represented by an unclassified ASV at day 33 which was mostly absent by day 39. Remarkably Cetobacterium species were conserved core affiliates throughout all fish from all treatments (Gallet et al. 2023).
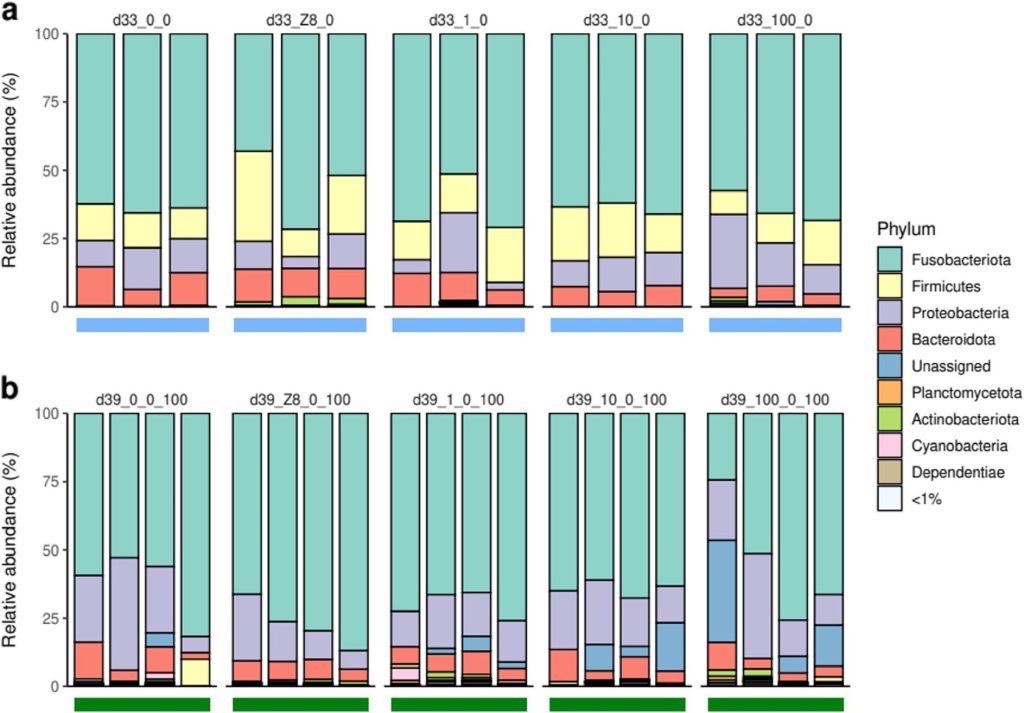
Fig 3. The relative abundances of the bacterial phyla of the fish gut microbiomes after a depuration phase from 28 to day 33, and a further six days of 100 µg l-1 Chla from day 33 to day 39. Image courtesy of Gallet et al. 2023 and the Creative Commons Licence 4.0. http://creativecommons.org/licenses/by/4.0/
There was little difference between the gut metabolomes of the fish from d28_Z8 and d28_1 and those at day 33, or from d28_Z8 and d28_10 and those of day 39. These triads differed from each other and the metabolomes of fish gut communities from other classes. Two Vibrio and two Reyranella species were negatively and positively associated with several metabolites. Liver metabolites were unaffected whilst muscle metabolites were similar at day 33, day 39, and in fish from d28_100, which were distinct from all other groups (Gallet et al. 2023).
Cetobacterium species were most abundant in gut, while ZOR0006 and Flavobacterium species were the most enriched in faeces, while environmental biofilms were monopolised by the genus Reyranella. All fish appeared healthy and survived, while blooms of Microcystis aeruginosa impact gut bacteria such as Firmicutes which are likely implicated in host metabolic processes. ZOR0006 were environmentally negligible and thus may be obligate intestinal microbiota of medaka fish. Cetobacterium species (Fusobacteria) are likely symbionts that manufacture 54 gut metabolic modules including one for vitamin B12 biosynthesis, while ZOR0006 utilise 15 conventional intestinal metabolites. Carbohydrate exploitation appears central to the metabolism of fish intestinal communities, while exceeding a bloom threshold seems to cause gut populations to shift. Reyranella species may be transitory gut residents whereas the genera Aeromonas, Flavobacterium, and Shewanella may be opportunistic pathogens or common core affiliates (Gallet et al. 2023).
Cyanopeptides like aerucyclamides and bacteriocins are potent antimicrobials and cytotoxins (Riley & Wertz 2002; Martins & Vasconcelos 2015). A change in microbial community structure does not necessarily spell disfunction especially with raised operational redundancy, however the metabolome shifted which may reflect host adaptive or immunological responses. Nevertheless there was a strong correlation between the microbial community compositions and the emergence of metabolites, where muscle variations were noted yet they were trivial compared to those of the gut (Gallet et al. 2023).
Remarkably, the growth medium modified prokaryotic gut communities and thus analogous effects are likely to arise within eutrophic fish-only recirculating systems or sea cage mariculture. Next we expand and consolidate our comprehension in part VIII as we delve deeper into pathobiome development.
References
Adams, J. (2018) Interesting Ecology Shift of Blacktail and Orangeface Butterflyfish. ReefBuilders.com. https://reefbuilders.com/2018/12/18/blacktail-and-redface-butterflyfish/
Akhter, N., Wu, B., Memon, A., M. & Mohsin, M. (2015) Probiotics and prebiotics associated with aquaculture: a review. Fish Shellfish Immunol. 45, 733-741. https://www.doi.org/10.1016/j.fsi.2015.05.038
Amann, R., I., Ludwig, W. & Schleifer, K., H. (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143-169.
Andreoni, F. & Magnani, M. (2014) Photobacteriosis: Prevention and Diagnosis. Journal of immunology research. 2014,.
Aslett, C., G. (2023a) Coral Immunity Part I. https://www.reefranch.co.uk/
Aslett, C., G. (2023b) Holosystemics Part IV: Dysbiosis and the Microscopic Coral Alliance. https://www.reefranch.co.uk/
Aslett, C., G. (2023c) Fish Immunostimulation. https://www.reefranch.co.uk/
Aslett, C., G. (2024) The Complete Reef Aquarist, for hobbyists, the trade and academics – A Conservation Manual. Reef Ranch Publishing Ltd, Leeds, West Yorkshire, UK.
Austin, B. & Austin, D., A. (2016) Bacterial fish pathogens, 6th ed. Springer, Stirling, UK
Austin, B. (1982) Taxonomy of bacteria isolated from a coastal, marine fish-rearing unit. J. Appl. Bacteriol. 53, 253-268. https://www.doi.org/10.1111/j.1365-2672.1982.tb04684.x
Bass, D., Stentiford. G., D., Wang, H-C., Koskella, B. & Tyler, C., R. (2019) The pathobiome in animal and plant diseases. Trends Ecol Evol. 34(11), 996-1008.
Bell, G., R., Hoskins, G., E. & Hodgkiss, W. (1971) Aspects of the characterization, identification, and ecology of the bacterial flora associated with the surface of stream-incubating Pacific salmon (Oncorhynchus) eggs. J. Fish. Board Can. 28, 1511-1525. https://www.doi.org/10.1139/f71-232
Birrell, C., Mccook, L., Willis, B. & Diaz-Pulido, G. (2008) Effects of benthic algae on the replenishment of corals and the implications for the resilience of coral reefs. Oceanography and marine biology. 46,.
Blazer, V., Phillips, S. & Pendleton, E. (2016) Fish Health, Fungal Infections, and Pfiesteria: The Role of the U.S. Geological Survey. U.S. Geological Survey Fact Sheet. pp 114-198.
Boilard, A., Dubé, C., E., Gruet, C., Mercière, A., Hernandez-Agreda, A. & Derome, N. (2020) Defining Coral Bleaching as a Microbial Dysbiosis within the Coral Holobiont. Microorganisms. 8, 1682. https://doi.org/10.3390/microorganisms8111682
Bone, Q., Marshall, N., B. & Blaxter, J., H., S. (1995) Biology of Fishes. Glasgow: Blackie Academic & Professional. https://www.doi.org/10.1007/978-1-4615-2664-3
Bozzi, D., Rasmussen, J., A., Carøe, C., et al. (2021) Salmon gut microbiota correlates with disease infection status: potential for monitoring health in farmed animals. Anim Microbiome. 3(1), 1-17.
Brandley, B., K. & Schnaar, R., L. (1986) Cell-Surface Carbohydrates in Cell Recognition and Response. J Leukoc Biol 40, 97–111. https://doi.org/10.1002/jlb.40.1.97
Brown, R., Moore, L., Mani, A., Patel, S., Salinas, I. (2021) Effects of ploidy and salmonid alphavirus infection on the skin and gill microbiome of Atlantic salmon (Salmo salar). PLoS One. 16(2), e0243684.
Cámara-Ruiz, M., Cerezo, I., M., Guardiola, F., A. et al. (2021) Alteration of the immune response and the microbiota of the skin during a natural infection by Vibrio harveyi in European seabass (Dicentrarchus labrax). Microorganisms. 9(5), 964.
Carnevali, O., Maradonna, F. & Gioacchini, G. (2017) Integrated control of fish metabolism, wellbeing and reproduction: the role of probiotic. Aquaculture 472, 144-155. https://www.doi.org/10.1016/j.aquaculture.2016.03.037
Cerezuela, R., Meseguer, J. & Esteban, M. (2011) Current knowledge in synbiotic use for fish aquaculture: a review. J. Aquac. Res. Dev. 1, 1-7.
Cherrak, Y., Flaugnatti, N., Durand, E., Journet, L. & Cascales, E. (2019) Structure and Activity of the Type VI Secretion System. Microbiology spectrum, 7(4), 1.
Colorni, A., Avtalion, R., Knibb, W., Berger, E., J., Colorni, B. & Timan, B. (1998) Histopathology of sea bass (Dicentrarchus labrax) experimentally infected with Mycobacterium marinum and treated with streptomycin and garlic (Allium sativum) extract. Aquaculture. 160, 1-17.
Cordero, H., Guardiola, F., A., Tapia-Paniagua, S., T., Cuesta, A., Meseguer, J., Balebona, M., C., et al. (2015) Modulation of immunity and gut microbiota after dietary administration of alginate encapsulated Shewanella putrefaciens Pdp11 to gilthead seabream (Sparus aurata L.). Fish Shellfish Immunol. 45, 608-618. https://www.doi.org/10.1016/j.fsi.2015.05.010
Das, P., Mandal, S., Khan, A., Manna, S., K. & Ghosh, K. (2014) Distribution of extracellular enzyme-producing bacteria in the digestive tracts of 4 brackish water fish species. Turk. J. Zool. 38, 79-88. https://www.doi.org/10.3906/zoo-1205-3
Delcroix, J., Gatesoupe, F., J., Desbruyères, E., Huelvan, C., Le Delliou, H., Le Gall, M., M., Quazuguel, P., et al. (2015) The effects of dietary marine protein hydrolysates on the development of sea bass larvae, Dicentrarchus labrax, and associated microbiota. Aquac. Nutr. 21, 98-104. https://www.doi.org/10.1111/anu.12139
Duperron, S., Halary, S., Habiballah, M., Gallet, A., Huet, H., Duval, C. et al. (2019) Response of fish gut microbiota to toxin-containing cyanobacterial extracts: a microcosm study on the Medaka (Oryzias latipes). Environ SciTechnol Lett. 6, 341–7.
Egerton, S., Culloty, S., Whooley, J., Stanton, C. & Ross, R., P. (2018) The Gut Microbiota of Marine Fish. Frontiers in Microbiology. 9(873),. https://www.frontiersin.org/articles/https://doi.org/10.3389/fmicb.2018.00873/full
Elhassan, Y., Philp, A. & Lavery, G. (2017) Targeting NAD+ in Metabolic Disease: New Insights Into an Old Molecule. Journal of the Endocrine Society. 1(7), 816-835.
Estruch, G., Collado, M., Peñaranda, D., Vidal, A., T., Cerdá, M., J., Martínez, G., P., et al. (2015) Impact of fishmeal replacement in diets for gilthead sea bream (Sparus aurata) on the gastrointestinal microbiota determined by pyrosequencing the 16S rRNA gene. PLoS One 10, e0136389. https://www.doi.org/10.1371/journal.pone.0136389
Flerova, E. & Balabanova, L. (2013) Ultrastructure of granulocytes of teleost fish (Salmoniformes, Cypriniformes, Perciformes). Journal of Evolutionary Biochemistry and Physiology. 49(2), 223-233.
Fournier, V., Huelvan, C. & Desbruyeres, E. (2004) Incorporation of a mixture of plant feedstuffs as substitute for fish meal in diets of juvenile turbot (Psetta maxima). Aquaculture 236, 451-465. https://www.doi.org/10.1016/j.aquaculture.2004. 01.035
Gallet, A., Halary, S., Duval, C., Huet, H., Duperron, S. & Marie, B. (2023) Disruption of fish gut microbiota composition and holobiont’s metabolome during a simulated Microcystis aeruginosa (Cyanobacteria) bloom. Microbiome. 11, 108. https://doi.org/10.1186/s40168-023-01558-2
Ghanbari, M., Kneifel, W. & Domig, K., J. (2015) A new view of the fish gut microbiome: advances from next-generation sequencing. Aquaculture 448, 464-475. https://www.doi.org/10.1016/j.aquaculture.2015.06.033
Giatsis, C., Sipkema, D., Smidt, H. et al. (2015) The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci Rep. 5(1), 1-15.
Green, T., J., Smullen, R. & Barnes, A., C. (2013) Dietary soybean protein concentrate-induced intestinal disorder in marine farmed Atlantic salmon, Salmo salar is associated with alterations in gut microbiota. Vet. Microbiol. 166, 286-292. https://www.doi.org/10.1016/j.vetmic.2013.05.009
Hansen, G. & Olafsen, J. (1999) Bacterial interactions in early life stages of marine cold water fish. Microb. Ecol. 38, 1-26. https://www.doi.org/10.1007/s002489900158
Hlongwane, P., Mungra, N., Madheswaran, S., Akinrinmade, O., A., Chetty, S. & Barth, S. (2018) Human Granzyme B Based Targeted Cytolytic Fusion Proteins. Biomedicines. 6(2), 72.
Ho, J. & Kim, I. (2001) New species of Hatschekia Poche, 1902 (Copepoda: Hatschekiidae) parasitic on marine fishes of Kuwait. Syst Parasitol. 49, 73-79.
Hooper, L., V. & Gordon, J., I. (2001) Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11, 1R-10R. https://doi.org/10.1093/glycob/11.2.1R
Hovanec, T. (2019) Dr. How to harness bacteria to cycle your saltwater tank quickly! | MACNA 2019. BrsTV. https://www.youtube.com/watch?v=zDI7sxqC-ss
James, A., G. (1988) Are clupeid microphagists herbivorous or omnivorous? A review of the diets of some commercially important clupeids. S. Afr. J. Mar. Sci. 7, 161-177. https://www.doi.org/10.2989/025776188784379017
Kapoor, B. & Khawna, B. (1993) The potential spectrum of the gut in teleost fishes. Adv. Fish Res. 1, 221-226.
Karachle, P., K. & Stergiou, K., I. (2010) Gut length for several marine fish: relationships with body length and trophic implications. Mar. Biodivers. Rec. 3, e106. https:///www.doi.org/10.1017/S1755267210000904
Kelly, C. & Salinas, I. (2017) Under pressure: Interactions between commensal microbiota and the teleost immune system. Front. Immunol. 8, 1.
Kim, B.,-R., Shin. J., Guevarra, R., B., et al. (2017) Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol. 27(12), 2089-2093.
Kim, J. & Lee, J., L. (2017) Correlation of Total Bacterial and Vibrio spp. Populations between Fish and Water in the Aquaculture System. Frontiers in Marine Science. 4,.
Kiron, V. (2012) Fish immune system and its nutritional modulation for preventive health care. Anim. Feed Sci. Technol. 173, 111-133. https://www.doi.org/10.1016/j.anifeedsci.2011.12.015
Kline, D., I., Kuntz, N., M., Breitbart, M., Knowlton, N. & Rohwer, F. (2006) Role of elevated organic carbon levels and microbial activity in coral mortality. Marine Ecology Progress Series. 314, 119-125.
Langille, M., G., Zaneveld, J., Caporaso, J., G. et al. (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 31(9), 814-821.
Larsen, A., M. (2014) Studies on the Microbiota of Fishes and the Factors Influencing Their Composition. Auburn, AL: Auburn University.
Lasica, A., Ksiazek, M., Madej, M. & Potempa, J. (2017) The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function. Frontiers in Cellular and Infection Microbiology. 7,.
Legrand, T., P., R., A., Catalano, S., R., Wos-Oxley, M., L., Stephens, F., Landos, M., Bansemer, M., S., Stone, D., A., J., Qin, J., G. & Oxley, A., P., A. (2018) The Inner Workings of the Outer Surface: Skin and Gill Microbiota as Indicators of Changing Gut Health in Yellowtail Kingfish. Front. Microbiol. 8:2664. https://www.doi.org/10.3389/fmicb.2017.02664
Lesel, R., De La Noüe, J. & Choubert, G. (1989) Fecal bacterial flora of rainbow trout under antibiotic treatment: effect of the number of pyloric caeca and the lipid content of food. Aquaculture: A Biotechnology in Progress, Vol. 1. De Pauw, N., Jaspers, E., Ackefors, H. & Wilkins, N. (eds.). Bredene: European Aquaculture Society, 592.
Li, P. & Gatlin, D., M. (2003) Evaluation of brewers yeast (Saccharomyces cerevisiae) as a feed supplement for hybrid striped bass (Morone chrysopsX—M. saxatilis). Aquaculture. 219(1), 681-692.
Li, T., Li, H., Gatesoupe, F-J. et al. (2017) Bacterial signatures of ‘red-operculum’ disease in the gut of crucian carp (Carassius auratus). Microb Ecol. 74(3), 510-521.
Liu, Q., Lai, Z., Gao, Y. et al. (2021) Connection between the gut microbiota of largemouth bass (Micropterus salmoides) and microbiota of the pond culture environment. Microorganisms. 9(8), 1770.
Llewellyn, M., Leadbeater, S., Garcia, C. et al. (2017) Parasitism perturbs the mucosal microbiome of Atlantic Salmon. Sci Rep. 7(1), 1-10.
Llewellyn, M., S., Boutin, S., Hoseinifar, S., H. & Derome, N. (2018) Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front Microbiol. 2014, 5. http://journal.frontiersin.org/article/10.3389/fmicb.2014.00207/abstract
Lobo, C., Moreno-Ventas, X., Tapia-Paniagua, S., Rodríguez, C., Moriñigo, M., A. & de La Banda, I., G. (2014) Dietary probiotic supplementation (Shewanella putrefaciens Pdp11) modulates gut microbiota and promotes growth and condition in Senegalese sole larviculture. Fish Physiol. Biochem. 40, 295-309. https://www.doi.org/10.1007/s10695-013-9844-0
Lom, J. & Corliss, J. (1971) Morphogenesis and Cortical Ultrastructure of Brooklynella hostilis, a Dysteriid Ciliate Ectoparasitic on Marine Fishes. The Journal of Eukaryotic Microbiology. 18(2), 261-281.
López Nadal, A., Peggs, D., Wiegertjes, G., F. & Brugman, S. (2018) Exposure to Antibiotics Affects Saponin ImmersionInduced Immune Stimulation and Shift in Microbial Composition in Zebrash Larvae. Front Microbiol 9, 2588. https://doi.org/10.3389/fmicb.2018.02588
López-Dóriga, M., Barnes, A., dos Santos, N. & Ellis, A. (2000) Invasion of fish epithelial cells by Photobacterium damselae subsp. piscicida: evidence for receptor specificity, and effect of capsule and serum. Microbiology. 146(1), 21-30.
Lorgen-Ritchie, M., Clarkson, M., Chalmers, L., Taylor, J., F., Migaud, H. & Martin, S., A. M. (2021) A temporally dynamic gut microbiome in Atlantic salmon during freshwater recirculating aquaculture system (RAS) production and post-seawater transfer. Front. Mar. Sci. 8, 869. https://doi.org/10.3389/fmars.2021.711797
Lorgen-Ritchie, M., Webster, T., U., McMurtrie, J., Bass, D., Tyler, C., R., Rowley, A. & Martin, S., A., M. (2023) Microbiomes in the context of developing sustainable intensified aquaculture. Frontiers in Microbiology. 14, 1200997.
Ma, C., Chen, C., Jia, L., He, X. & Zhang, B. (2019) Comparison of the intestinal microbiota composition and function in healthy and diseased Yunlong Grouper. AMB Express. 9(1), 1-11.
Martins, J. & Vasconcelos, V. (2015) Cyanobactins from cyanobacteria: current genetic and chemical state of knowledge. Mar Drugs. 13, 6910-6946.
Mashoof, S. & Criscitiello, M., F. (2016) Fish Immunoglobulins. Biology. 5(4), 45.
McBride, M., J. & Nakane, D. (2015) Flavobacterium gliding motility and the type IX secretion system. Curr. Opin. Microbiol. 28, 72-77.
Merrifield, D., L. & Rodiles, A. (2015) The fish microbiome and its interactions with mucosal tissues. Mucosal Health in Aquaculture. Peatman, E. (ed.) Academic Press, San Diego, CA. pp 273-295.
Mihalitsis, M. & Bellwood, D. (2017) A morphological and functional basis for maximum prey size in piscivorous fishes. PLoS ONE. 12(9),.
Miyake, S., Ngugi, D. K. & Stingl, U. (2015) Diet strongly influences the gut microbiota of surgeonfishes. Mol. Ecol. 24, 656-672. https://www.doi.org/10.1111/mec.13050
Miyake, S., Soh, M., Azman, M., N., Ngoh, S., Y., Orbán, L., Seedorf, H. (2020) Insights into the microbiome of farmed Asian sea bass (Lates cal-carifer) with symptoms of tenacibaculosis and description of Tenacibaculum singaporense sp. nov. Antonie Van Leeuwenhoek. 113(6), 737-752.
Monroig, Ó., Tocher, D., R. & Navarro, J., C. (2013) Biosynthesis of polyunsaturated fatty acids in marine invertebrates: recent advances in molecular mechanisms. Mar. Drugs 11, 3998-4018. https://www.doi.org/10.3390/md11103998
Moon, D. (2021) Boosting NAD+ to Reverse Aging? Overview of NR and NMN. GeneticLifeHacks.com. https://www.geneticlifehacks.com/nad-reversing-aging-overview-of-nr-and-nmn/
Mouchet, M., A., Bouvier, C., Bouvier, T., Troussellier, M., Escalas, A. & Mouillot, D. (2012) Genetic difference but functional similarity among fish gut bacterial communities through molecular and biochemical fingerprints. FEMS Microbiol. Ecol. 79, 568-580. https://www.doi.org/10.1111/j.1574-6941.2011. 01241.x
Mougin, J. & Joyce, A. (2022) Reviews in Aquaculture. Fish disease prevention via microbial dysbiosis-associated biomarkers in aquaculture. 15, 579-594. https://doi.org/10.1111/raq.12745
Muñoz-Baquero, M., Lorenzo-Rebenaque, L., Garc´ıa-Va´zquez, F., A., Garc´ıa-Pa´rraga, D., Mart´ınez-Priego, L., De Marco-Romero, G., Gala´n-Vendrell, I., D’Auria, G. & Marco-Jime´nez, F. (2023) Unveiling Microbiome Signature In Inner Body Fluids: Comparison Between Wild And Aquarium Small-Spotted Catshark (Scyliorhinus canicular). Frontiers in Marine Science. https://www.doi.org/10.3389/fmars.2023.1151119
Murdoch, C., C. & Rawls, J., F. (2019) Commensal microbiota regulate vertebrate innate immunity-insights from the zebrafish. Front. Immunol. 10:2100. https://doi.org/10.3389/fimmu.2019.02100
Navarrete, P., Espejo, R. T. & Romero, J. (2009) Molecular analysis of microbiota along the digestive tract of juvenile Atlantic salmon (Salmo salar L.). Microb. Ecol. 57, 550-561. https://www.doi.org/10.1007/s00248-008-9448-x
Naya-Català, F., Piazzon, M., C., Calduch-Giner, J. A., Sitjà-Bobadilla, A. & Pérez-Sánchez, J. (2022) Diet and host genetics drive the bacterial and fungal intestinal metatranscriptome of Gilthead Sea bream. Front. Microbiol. 13, 883738. https://doi.org/10.3389/fmicb.2022.883738
Nayak, S., K. (2010) Role of gastrointestinal microbiota in fish. Aquac. Res. 41, 1553-1573. https://www.doi.org/10.1111/j.1365-2109.2010.02546.x
Neuman, C., Hatje, E., Zarkasi, K. Z., Smullen, R., Bowman, J., P. & Katouli, M. (2016) The effect of diet and environmental temperature on the faecal microbiota of farmed Tasmanian Atlantic Salmon (Salmo salar L.). Aquac. Res. 47, 660-672. https://www.doi.org/10.1111/are.12522
Noga, E., J. (2010) Fish Disease. Diagnosis and Treatment. Second Edition. Wiley-Blackwell, John Wiley & Sons Inc. p 139.
Oliva-Teles, A. & Goncalves, P. (2001) Partial replacement of fishmeal by brewers yeast (Saccaromyces cerevisae) in diets for sea bass (Dicentrarchus labrax) juveniles. Aquaculture. 202(3), 269-278.
Orlandi, I., Alberghina, L. & Vai, M. (2020) Nicotinamide, Nicotinamide Riboside and Nicotinic Acid-Emerging Roles in Replicative and Chronological Aging in Yeast. Biomolecules. 10(4), 604.
Ortiz-Estrada, A., M., Gollas-Galván, T., Martínez-Cordova, L., R. & Martínez-Porchas, M. (2019) Predictive functional profiles using metagenomic 16S rRNA data: a novel approach to understanding the microbial ecology of aquaculture systems. Rev Aquacult. 11(1), 234-245.
Perdiguero, P., Martin-Martin, A., Benedicenti, O., Diaz-Rosales, P., Morel, E., Munoz-Atienza, E., Garcia-Flores, M., Simon, R., Soleto, I., Cerutti, A. & Tafalla, C. (2019) Teleost IgD+Ig- B Cells Mount Clonally Expanded and Mildly Mutated Intestinal IgD Responses in the Absence of Lymphoid Follicles. Cell Reports. 29(13), 4223-4235.
Pérez-Pascual, D., Lunazzi, A., Magdelenat, G., Rouy, Z., Roulet, A., Lopez-Roques, C., Larocque, R., Barbeyron, T., Gobet, A., Michel, G., Bernardet, J. & Duchaud, E. (2017) The Complete Genome Sequence of the Fish Pathogen Tenacibaculum maritimum Provides Insights into Virulence Mechanisms. Frontiers in Microbiology. 8,.
Reid, K., M., Patel, S., Robinson, A., J., et al. (2017) Salmonid alphavirus infection causes skin dysbiosis in Atlantic salmon (Salmo salar L.) post-smolts. PLoS One. 12(3):e0172856.
Richardson, L., L., Sekar, R., Myers, J., L., Gantar, M., Voss, J., D., Kaczmarsky, L., Remily, E., R., Boyer, G., L. & Zimba, P., V. (2007) The presence of the cyanobacterial toxin microcystin in black band disease of corals. FEMS Microbiology Letters. 272(2), 182-187.
Riley, M., A. & Wertz, J., E. (2002) Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 84, 357-364.
Ringø, E., Lødemel, J., B., Myklebust, R., Jensen, L., Lund, V., Mayhew, T., M., et al. (2002) The effects of soybean, linseed and marine oils on aerobic gut microbiota of Arctic charr Salvelinus alpinus L. before and after challenge with Aeromonas salmonicida ssp. salmonicida. Aquac. Res. 33, 591-606. https://www.doi.org/10.1046/j.1365-2109.2002.00697.x
Ringø, E., Zhou, Z., Vecino, J., L., G., Wadsworth, S., Romero, J., Krogdahl, Å., et al. (2016) Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac. Nutr. 22, 219-282. https://www.doi.oeg/10.1111/anu.12346
Ritchie, K., B. & Smith, G., W. (1995) Preferential carbon utilization by surface bacterial communities from water mass, normal, and white-band diseased Acropora cervicornis. Mol Mar Biol Biotechnol. 4, 345-354.
Rohde, K. (1932) Ciliophora (ciliates). Marine Parasitology. Rohde, K. (ed.). CSIRO Publishing, Clayton, Australia.
Rombout, J., H., Abelli, L., Picchietti, S., Scapigliati, G. & Kiron, V. (2011) Teleost intestinal immunology. Fish Shellfish Immunol. 31, 616-626. https://www.doi.org/10.1016/j.fsi. 2010.09.001
Rosado, D., Pérez-Losada, M., Pereira, A., Severino, R. & Xavier, R. (2021) Effects of aging on the skin and gill microbiota of farmed seabass and seabream. Anim Microbiome. 3(1), 1-14.
Rosado, D., Pérez-Losada, M., Severino, R. & Xavier, R. (2022) Monitoring infection and antibiotic treatment in the skin microbiota of farmed European seabass (Dicentrarchus Labrax) fingerlings. Microb Ecol. 83(3), 789-797.
Rosado, D., Perez-Losada, M., Severino, R., Cable, J. & Xavier, R. (2019b) Characterization of the skin and gill microbiomes of the farmed seabass (Dicentrarchus labrax) and seabream (Sparus aurata). Aquaculture. 500, 57-64.
Rosado, D., Xavier, R., Severino, R., Tavares, F., Cable, J. & Pérez-Losada, M. (2019a) Effects of disease, antibiotic treatment and recovery trajectory on the microbiome of farmed seabass (Dicentrarchus labrax). Sci Rep. 9(1), 1-11.
Rozas-Serri, M. (2019) Gill diseases in marine salmon aquaculture with an emphasis on amoebic gill disease. CAB Reviews Perspectives in Agriculture Veterinary Science Nutrition and Natural Resources. 14, 1-15.
Santoro, E., P., Borges, R., M., Espinoza, J., L., Freire, M., Messias, C., S., M., A., Villela, H., D., M., Pereira, L., M., Vilela, C., L., S., Rosado, J., G., Cardoso, P., M., Rosado, P., M., Assis, J., M., Duarte, G., A., S., Perna, G., Rosado, A., S., Macrae, A., Dupont, C., L., Nelson, K., E., Sweet, M., J., Voolstra, C., R. & Peixoto, R., S. (2021) Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Science advances. 7(33), 3088.
Santos, F., F., Yamamoto, D., Abe, C., M., Bryant, J., A., Hernandes, R., T., Kitamura, F., C., Castro, F., S., Valiatti, T., B., Piazza, R., Elias, W., P., Henderson, I., R. & Gomes, T. (2019) The Type III Secretion System (T3SS)-Translocon of Atypical Enteropathogenic Escherichia coli (aEPEC) Can Mediate Adherence. Frontiers in microbiology. 10, 1527.
Scapigliati, G., Fausto, A., M. & Picchietti, S. (2018) Fish Lymphocytes: An Evolutionary Equivalent of Mammalian Innate-Like Lymphocytes? Frontiers in immunology. 9, 971.
Segata, N., Izard, J., Waldron, L., et al. (2011) Metagenomic biomarker discovery and explanation. Genome Biol. 12(6), 1-18.
Shabir, U., Ali, S., Magray, A., Ganai, B., Firdous, P., Hassan, T. & Nazir, R. (2018) Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microbial pathogenesis. 114, 50-56.
Sila, A., Nedjar-Arroume, N., Hedhili, K., Chataigné, G., Balti, R., Nasri, M., et al. (2014) Antibacterial peptides from barbel muscle protein hydrolysates: activity against some pathogenic bacteria. LWT Food Sci. Technol. 55, 183-188. https://www.doi.org/10.1016/j.lwt.2013.07.021
Siriyappagouder, P., Galindo-Villegas, J., Dhanasiri, A., K., S., Zhang, Q., Mulero, V., Kiron, V. et al. (2020) Pseudozyma priming influences expression of genes involved in metabolic pathways and immunity in zebrafish larvae. Front. Immunol. 11, 987. https://doi.org/10.3389/fimmu.200.00978
Siriyappagouder, P., Kiron, V., Lokesh, J., Rajeish, M., Kopp, M. & Fernandes, J. (2018) The intestinal mycobiota in wild zebrafish comprises mainly Dothideomycetes while Saccharomycetes predominate in their laboratory-reared counterparts. Front. Microbiol. 9, 387. https://doi.org/10.3389/fmicb.2018.00387
Sitjà-Bobadilla, A., Gil-Solsona, R., Estensoro, I., Piazzon, M., Martos-Sitcha, J., Picard-Sánchez, A., Fuentes, J., Sancho, J., Calduch-Giner, J., Hernández, F. & Pérez-Sánchez, J. (2019) Disruption of gut integrity and permeability contributes to enteritis in a fish-parasite model: a story told from serum metabolomics. Parasites & Vectors. 12,.
Skrodenyte-ArbaCIauskiene, V. (2007) Enzymatic activity of intestinal bacteria in roach Rutilus rutilus L. Fish. Sci. 73, 964-966. https://www.doi.org/10.1111/j.1444-2906.2007. 01421.x
Slinger, J., Adams, M., B., Stratford, C., N., Rigby, M. & Wynne, J., W. (2021) The effect of antimicrobial treatment upon the gill bacteriome of Atlantic salmon (Salmo salar L.) and progression of amoebic gill disease (AGD) in vivo. Microorganisms. 9, 987. https://doi.org/10.3390/microorganisms9050987
Spanova, M. & Daum, G. (2011) Squalene–biochemistry, molecular biology, process biotechnology, and applications. Eur J Lipid Sci Technol. 113(11), 1299-1320.
Spor, A., Koren, O. & Ley, R. (2011) Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279-290. https://doi.org/10.1038/nrmicro2540
Stevens, C., E. & Hume, I., D. (2004) Comparative Physiology of the Vertebrate Digestive System. Cambridge: Cambridge University Press.
Stevens, J., Jackson, R., Olson, J. (2016) Bacteria associated with lionsh (Pterois volitans/miles complex) exhibit antibacterial activity against known fish pathogens. Mar Ecol Prog Ser 558, 167–180. https://doi.org/10.3354/meps11789
Sullam, K., E., Essinger, S., D., Lozupone, C., A., O’Connor, M., P., Rosen, G., L., Knight, R., et al. (2012) Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21, 3363-3378. https://www.doi.org/10.1111/j.1365-294X.2012.05552.x
Swiatecka, D., Markiewicz, L., H. & Wróblewska, B. (2012) Pea protein hydrolysate as a factor modulating the adhesion of bacteria to enterocytes, epithelial proliferation and cytokine secretion–an in vitro study. Cent. Eur. J. Immunol. 37, 227-231. https://www.doi.org/10.5114/ceji.2012.3079
Tapia-Paniagua, S., T., Ceballos-Francisco, D., Balebona, M., C., Esteban, M., A. & Moriñigo, M., A. (2018) Mucus glycosylation, immunity and bacterial microbiota associated to the skin of experimentally ulcered gilthead seabream (Sparus aurata). Fish Shellfish Immunol. 75, 381-390.
Thiéry, R., Cozien, J., Cabon, J., Lamour, F., Baud, M. & Schneemann, A. (2006) Induction of a Protective Immune Response against Viral Nervous Necrosis in the European Sea Bass Dicentrarchus labrax by Using Betanodavirus Virus-Like Particles. Journal of Virology. 80(20), 10201.
Torrecillas, S., Montero, D. & Izquierdo, M. (2014) Improved health and growth of fish fed mannan oligosaccharides: potential mode of action. Fish Shellfish Immunol. 36, 525-544. https://doi.org/10.1016/j.fsi.2013.12.029
Tretina, K., Park, E., S., Maminska, A. & MacMicking, J., D. (2019) Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. The Journal of Experimental Medicine. 216(3), 482-500.
Valero, Y., Saraiva‐Fraga, M., Costas, B. & Guardiola, F., A. (2019) Antimicrobial peptides from fish: beyond the fight against pathogens. Reviews in Aquaculture. 12, 224-253.
Vayssier-Taussat, M., Albina, E., Citti, C., et al. (2014) Shifting the paradigm from pathogens to pathobiome: new concepts in the light of metaomics. Front Cell Infect Microbiol. 4, 29.
Vestrum, R., I., Forberg, T., Luef, B., Bakke, I., Winge, P., Olsen, Y., et al. (2022) Commensal and opportunistic bacteria present in the microbiota in Atlantic cod (Gadus morhua) larvae differentially alter the hosts’ innate immune responses. Microorganisms. 10, 24. https://doi.org/10.3390/microorganisms10010024
Walke, J., B., et al. (2017) Dominance-function relationships in the amphibian skin microbiome. Environ. Microbiol. 19, 3387–3397.
Wang, S. & Loreau, M. (2014) Ecosystem stability in space: α, β and γ variability. Ecol Lett. 17(8), 891-901.
Webster, T., M., U., Rodriguez-Barreto, D., Consuegra, S. & Garcia de Leaniz, C. (2019) Cortisol-induced signatures of stress in the fish microbiome. bioRxiv:826503. https://doi.org/10.1101/826503
Wilson, J., M. & Castro, L., F., C. (2010) Morphological diversity of the gastrointestinal tract in fishes. Fish Physiol. 30, 1-55. https://www.doi.org/10.1016/S1546-5098(10)03001-3
Wilson, M. World Feeds Limited, 3b Coulman Street Industrial Estate, Thorne, Doncaster, DN8 5JS, United Kingdom.
Wu, J., Mao, C., Deng, Y., et al. (2019) Diversity and abundance of antibiotic resistance of bacteria during the seedling period in marine sh cage-culture areas of Hainan, China. Mar Pollut Bull 141:343–349. https://doi.org/10.1016/j.marpolbul.2019.02.069
Xavier, R., Pereira, A., Pagan, A., Hendrick, G., C., Nicholson, M., D., Rosado, D., Soares, M., C., Pérez-Losada, M., Sikkel, P., C. (2020) The effects of environment and ontogeny on the skin microbiome of two Stegastes damselfishes (Pomacentridae) from the eastern Caribbean Sea. Mar Biol. 167(7),1-12.
Xu, Z., Takizawa, F., Casadei, E., Shibasaki, Y., Ding, Y., Sauters, T., J., C., et al. (2020) Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol. 5, 3254. https://doi.org/10.1126/sciimmunol.aay3254
Yatsunenko, T., Rey, F., E., Manary, M., J., Trehan, I., Dominguez-Bello, M., G., Contreras, M., et al. (2012) Human gut microbiome viewed across age and geography. Nature 486, 222-227. https://www.doi.org/10.1038/nature11053
Yoon, J., Matsuo, Y., Matsuda, S., Adachi, K., Kasai, H. & Yokota, A. (2018) Rubritalea sabuli sp. nov., a carotenoid-and squalene-producing member of the family Verrucomicrobiaceae, isolated from marine sediment. Int J Syst Evol Microbiol. 58(4), 992-997.
Yoshimizu, M., Kimura, T. & Sakai, M. (1980) Microflora of the embryo and the fry of salmonids. Bull. Jpn. Soc. Sci. Fish. 46, 967-975. https://www.doi.org/10.2331/suisan.46.967
Yu, Y., Wang, Q., Huang, Z., Ding, L. & Xu, Z. (2020) Immunoglobulins, Mucosal Immunity and Vaccination in Teleost Fish. Frontiers in immunology. 11, 567941.
Zanefeld, J., R. et al. (2017) Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2, 17121.
Zarkasi, K., Z., Taylor, R., S., Abell, G., C., Tamplin, M., L., Glencross, B., D. & Bowman, J., P. (2016) Atlantic salmon (Salmo salar L.) gastrointestinal microbial community dynamics in relation to digesta properties and diet. Microb. Ecol. 71, 589-603. https://www.doi.org/10.1007/s00248-015-0728-y
Zeng, A., Tan, K., Gong, P. et al. (2020) Correlation of microbiota in the gut of fish species and water. 3 Biotech. 0(11), 1-10.
Zhang, L., Ni, C., Xu, W., Dai, T., Yang, D., Wang, Q., Zhang, Y. & Liu, Q. (2016) Intramacrophage Infection Reinforces the Virulence of Edwardsiella tarda. Journal of bacteriology. 198(10), 1534-1542.
Zhang, M., Shan, C., Tan, F., Limbu, S., M., Chen, L. & Du, Z. (2020) Gnotobiotic models: powerful tools for deeply understanding intestinal microbiota-host interactions in aquaculture. Aquaculture 517:734800. https://doi.org/10.1016/j.aquaculture.2019.734800
Zhang, X., Ding, L., Yu, Y. et al. (2018) The change of teleost skin commensal microbiota is associated with skin mucosal transcriptomic responses during parasitic infection by Ichthyophthirius multifillis. Front Immunol. 9, 2972.
Zhang, Y. & Gui, J. (2012) Molecular regulation of interferon antiviral response in fish. Developmental and comparative immunology. 38(2), 193-202.
Zhou, Z., Liu, Y., Shi, P., He, S., Yao, B. & Ringø, E. (2009) Molecular characterization of the autochthonous microbiota in the gastrointestinal tract of adult yellow grouper (Epinephelus awoara) cultured in cages. Aquaculture 286, 184-189. https://www.doi.org/10.1016/j.aquaculture.2008.10.002
Zhou, Z., Yao, B., Romero, J., Waines, P., Ringø, E., Emery, M., et al. (2014) Methodological approaches used to assess fish gastrointestinal communities. Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics, Merrifield, D. & Ringø, E. (eds.). John Wiley & Sons Ltd., Hoboken, NJ.


0 Comments