Our understanding of disease is evolving because many do not originate from the exploits of a primary pathogen, which do not merely interact with the host and its immune system, but with the multipartite holobiont and its immediate surroundings. Illnesses appear a consequence of a dynamic process whereby microeukaryotic and prokaryotic symbionts, commensals, and their viruses, initially attempt to outcompete and eradicate invading microorganisms by liberating antimicrobials and modifying their community compositions and metabolism. Cellular trauma that exceeds their restorative potential demands participation of adaptive and innate immunity. Host defense transforms the conditions within its microbiomes to curtail spread and purge disease-causing agents, which leads to transitory and intended states of dysbiosis. The pathobiont is the consequence of the host’s altered constituent physiology and its now hostile microniche (pathobiome), and its modified community and metabolic function (pathobolome) which we hitherto called disease (Bass et al. 2019).

Fig 1. A color variant of the percula clownfish (Amphiprion percula) stricken with Brooklynella cf. hostilis with a moribund posture comprising “clamped” fins. Image courtesy of Alex Cole ©, Meridian, Idaho, USA.
The term symbiome refers to a microniche and its microbial community including microeukaryotes, prokaryotes, and their viruses which may be benign, mutual- or antagon-istic, whereas skin-residing mutualists are often referred to as epi-symbionts. Interactions may be short or long lived, where pathobiome development is often gradual and advocates novel mitigating approaches. Investigations must consider abiotic and biotic contributions insofar as pathobiomes may arise from unfavorable conditions and invasions of environmental microbes (Bass et al. 2019), while commensals like Brooklynella hostilis may proliferate and aggressively erode gill and skin (Fig 1.; Rohde 1932; Lom & Corliss 1971; Noga 2010).

Fig 2. A post-mortem example of freshwater kuria labeo (Labeo gonius) exhibiting epizootic ulcerative syndrome (EUS) caused by the fungus-like chromist Aphanomyces invadans which is a disease epitomizing hyphae-derived host-specific aetiologies, site-specific ulcerations, petechial hemorrhages, and tail erosion. Nevertheless A. invadans is constrained to hyposaline or freshwater (Blazer, personal communication).
Fish pathogens include microorganisms from every kingdom where an established lesion is an ecological niche where a consortium of microorganisms exploit a range of nutrition, whereas abiotic environmental impacts and immune subversion may recrudesce dormant aetiological agents (cryptic pathogens; Bass et al. 2019). A consortium may act synergistically to intensify pathology, like the infamous coinfection of Atlantic menhaden with Pfiesteria species of dinoflagellate and the fungi-like chromist Aphanomyces invadans from 1996 to 1997 in Chesapeake Bay (Fig 2.; Blazer et al. 2016), yet equally, multiple contagions can prove aetiologically attenuating.
Bacterial quorum quenching degrades the agenda-orchestrating signaling autoinducers of other prokaryotes, or the complementary metabolites of a few symbiome or pathobiome affiliates may be shared (Bass et al. 2019). Several pathogens are recognized inhabitants of “healthy” microbiomes yet increases of host apoptosis or necrosis trigger their logarithmic growth and community destabilization beyond a predetermined threshold. Such disproportionate proliferation within a finite niche forces taxa extinction and a loss of diversity. Inter-symbiont horizontal gene transfer arms benign microbes with virulence factors and frequencies are elevated by stress (Bass et al. 2019).
The “Anna Karenina” principle proposes the microbial compositions of dysbiotic organisms vary more than those of “healthy” (Zanefeld et al. 2017, cited in Bass et al. 2019) however in stark contrast to many studies, a succession of limited microbial taxa and a dip in diversity has been associated with mitigation (Webster et al. 2019, cited in Lorgen-Ritchie et al. 2023). Certain prokaryotes confer resistance to specified disease-causing agents where these lineages were enriched in frog skin from pathogen-infested environments yet appeared nominally in pathogen-free milieus (Walke et al. 2017, cited in Bass et al. 2019). Probiotic manipulations are tangible goals for corals and fish and their immediate surroundings however we must exercise caution. The microbiomes of “healthy” corals are unaffected by beneficial microbes (BMCs; Santoro et al. 2021) yet attempting to exclude perceived pathogens from symbiomes is hazardous insofar as there are significant gaps in our understanding (Bass et al. 2019).
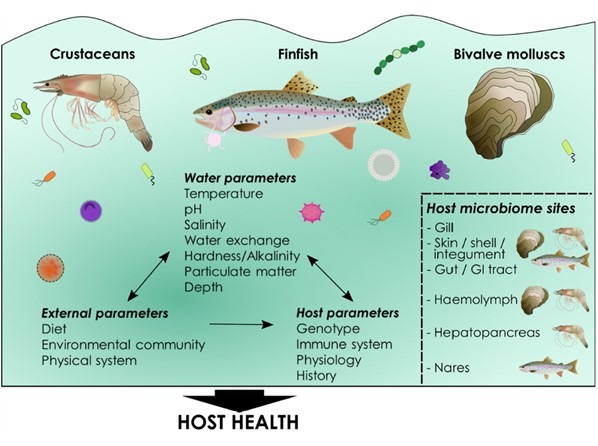
Fig 3. Holobiont/environmental dynamics. Image courtesy of Lorgen-Ritchie et al. 2023 and the Creative Commons Attribution Licence (CC BY). http://creativecommons.org/licenses/by/4.0/
The gut of zebrafish includes 15 fungi belonging to the phyla Ascomycota, Basidiomycota, and Zygomycota, yet the communities of wild and captive fish a dominated by contrasting kinds (Siriyappagouder et al. 2018), while fungi modulate the digestion, immunity, and hormones of gilthead seabream (Sparus aurata; Naya-Català et al. 2022). The communities of planktonic microbes in both recirculating and sea cage environments are shaped by abiotic and biotic factors such as the weather and husbandry in dynamic flux. Finfish fry adaptive and innate immunity are not primed and external and internal microbes do not colonize until after hatching and first feeds, where microbe-derived bacteriocins, fungicides, antimicrobial peptides (AMPs), viruses/bacteriophage, and immunity commence to structure microbial populations (Lorgen-Ritchie et al. 2023). Cell surface receptors encoded by cell germlines may play a central role in recruitment and winnowing which are likely tissue-specific, and akin to corals there is evidence of sophisticated “crosstalk” between microbes and host immunity (Kelly & Salinas 2017).
Unless infected vertically in gamete mucus, fish are only colonized by immunity-evading/resilient environmental microbes (Lorgen-Ritchie et al. 2023) whereas host genotype influences prokaryotic recognition, sifting, and the biochemical composition of its microclimates (Spor et al. 2011). Host genotype and epigenetic modification may have a profound effect on microbial communities which is impacted by cellular metabolites, while invertebrates can exhibit lasting transgenerational immunity. Microbiomes may be shaped in aquaculture using targeted environmental conditioning (Lorgen-Ritchie et al. 2023) inasmuch as the ratios of organics to inorganics determines heterotrophy versus autotrophy, where the former can overgrow biofilms or outcompete planktonic nitrifying autotrophs responsible for detoxification of ammonia (Hovanec, personal communication 2019). Water turnover, degas/regas, and purification has a direct impact on microbes which become microbiome affiliates, whereas the introduction of healthy animals can act as welcome inoculants. System and animal fitness are thus interrelated where function or malfunction are expedited on a sliding scale in dynamic equilibrium. Defining optimal conditions is challenging in aquaculture due to the variety of methodologies, while parameter manipulation remains child’s play in recirculating systems (Lorgen-Ritchie et al. 2023).
Pathogens such as Vibrio and Photobacterium species (Vibrionaceae) proliferate in response to increased temperature while circadian rhythm and thus homeostasis are influenced by photoperiod. Biofloc technology is used in recirculating aquaculture systems (RAS) to maintain healthy nitrifying communities which in turn supports livestock defense (Lorgen-Ritchie et al. 2023). Feeding the inhabitants of an ornamental tropical marine fish-only systems several times per day has an analogous effect. Heavy feeds are not uncommon in aquaculture yet they devastate reefs, while forms of sterilization may prove harmful, and biofouling exerts impacts (Lorgen-Ritchie et al. 2023). Detrital purging is thus important in all recirculating systems, however cleaning deep sand beds and “plenums” is risky.
A “healthy” microbiome must respond and adapt to varying conditions, support host homeostasis, and provide resilience to disease, where function and the metabolome is more vital than phylogeny. Compliant populations contribute to host phenotypic plasticity, whereas stress-mediated host cortisol impacts community dynamics and may be used as a biomarker (Lorgen-Ritchie et al. 2023).
Dysbiosis and rebiosis occur during smoltification of Atlantic salmon a week after holobionts transition from fresh- to salt-water, where contrasting populations are established by week four (Lorgen-Ritchie et al. 2021). Chemotherapeutic antimicrobial agents have resulted in microbiome destabilization and an escalation of amoebic gill disease (AGD; Slinger et al. 2021) caused by a complex and somewhat interchangeable consortium (Rozas-Serri 2019).
High-throughput amplicon sequencing has been used to profile microbial communities and metabolomes, however it is limited inasmuch as databases of microbiome affiliates are underpopulated, where many such gene sets are transcriptionally inactive or downregulated. Holistic“omic” approaches like metagenomics, metatranscriptomics, metaproteomics, and metabolomics combined into multi-omics and “hologenomics” attempt to elucidate and contextualise the interactions of cross-anatomy cross-environmental genes, their products, and those of the host, which rely upon extensive sampling and yet to be established bioinformatics. Innovative exploitation of hologenomics will likely highlight the contrasting and alike functional dynamics and redundancy of fish holobionts (Lorgen-Ritchie et al. 2023).
Gnotobiotic animals are reared sterile in a sterile environment which may be used to clarify the impact of a single microbe or cohort (Zhang et al. 2020) where studies have elucidated how gut microbiota exert vital immunological and morphological impacts in the intestines of developing cod (Gadus morhua; Vestrum et al. 2022). Waterborne bacteria inoculated into the gut of germ-free zebrafish expressed 212 genes 59 of which were also detected in mouse intestines which is redolent of evolutionary conservation; however teleostean recruitment is more selective than murine (Murdoch & Rawls 2019). 59 genes were also differentially expressed in the gut of sterile zebrafish infected with Pseudozyma species of yeast exemplifying the proficiency of interkingdom taxa in metabolism and immunity, yet no changes were observed in exposed conventionally reared fish (Siriyappagouder et al. 2020). The phylogenetic lineages of fish gut inoculums became microbiome affiliates, yet their community compositions consistently aligned with the species of their host. These studies suggest such strategies may be safely used for restorative therapies (Lorgen-Ritchie et al. 2023).
Recombinant, “knockout” and “knock-in” genetic approaches whereby organisms are genetically modified unravel the intricacies of microbial consortium dynamics. Genetic modification of rainbow trout (Oncorhynchus mykiss) limited their mucosal titres of secretory immunoglobulin T (sIgT) which are crucial for pathogen suppression and microbiome stability. sIgT– mutants exhibited a negligible IgM response, were dysbiotic, and experienced cellular trauma and inflammation compared with wildtype (Xu et al. 2020). Finfish clones offer consistent microbiomes for comparative studies, whereas the renaissance of in vitro bacterial culture provides opportunities to study and characterize otherwise rare prokaryotic consortia which populates and consolidates metaproteomic, metagenomic, and metabolomic databases (Lorgen-Ritchie et al. 2023).
Manipulations exploiting beneficial microbes for fish (BMFs) must be cost effective and comprise stable species and strains, that may be prominent key taxa implicated in disease mitigation or immunity, homeostatic endurance, or rebiosis which will likely be a handful of taxa or discrete subspecies. Cyst-forming heterotrophic microbes are stable in bottles yet they outcompete and overgrow autotrophic nitrifiers which may result in detectable nitrite (Hovanec, personal communication 2019).
Probiotics are diets comprising live microbes that elicit competitive exclusion, while prebiotics contain vital nutrients know to stimulate the proliferation of “good” bacteria. These therapies promote synergy as combined synbiotics (Lorgen-Ritchie et al. 2023) whereas fish immunostimulation using household and commercial products is explored in Aslett 2023c.
Therapies should be timed to optimize efficacy throughout critical developmental stages such as hatching, larval rearing, saltwater acclimation, or during heat and/or hypoxia-induced stress or dysbiosis, which warrants further investigation. Pro- and pre-biotics are conventionally dietary, while manipulative cultures can be applied directly to water in recirculating systems to transform mucosal populations. However this discipline is in its infancy and random inoculations of bacterial cultures will most likely prove harmful, whereas studies of fungal, viral and microeukaryotic symbionts present novel remedial approaches. Consortium-specific AMPs represent an innovative therapy which should strengthen commensals/mutualists whilst precluding pathogens and resistance, where function rather than phylogeny appear decisive (Lorgen-Ritchie et al. 2023).
Next we examine the dynamics of the skin mucosal microbiome of juvenile seabass throughout challenges with wild pathogens thanks to the discerning research of Rosado and allies from 2022.
Editor’s note: the final articles in this series are also available at: https:www.reefranch.co.uk/
References
Adams, J. (2018) Interesting Ecology Shift of Blacktail and Orangeface Butterflyfish. ReefBuilders.com. https://reefbuilders.com/2018/12/18/blacktail-and-redface-butterflyfish/
Akhter, N., Wu, B., Memon, A., M. & Mohsin, M. (2015) Probiotics and prebiotics associated with aquaculture: a review. Fish Shellfish Immunol. 45, 733-741. https://www.doi.org/10.1016/j.fsi.2015.05.038
Amann, R., I., Ludwig, W. & Schleifer, K., H. (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143-169.
Andreoni, F. & Magnani, M. (2014) Photobacteriosis: Prevention and Diagnosis. Journal of immunology research. 2014,.
Aslett, C., G. (2023a) Coral Immunity Part I. https://www.reefranch.co.uk/
Aslett, C., G. (2023b) Holosystemics Part IV: Dysbiosis and the Microscopic Coral Alliance. https://www.reefranch.co.uk/
Aslett, C., G. (2023c) Fish Immunostimulation. https://www.reefranch.co.uk/
Aslett, C., G. (2024) The Complete Reef Aquarist, for hobbyists, the trade and academics – A Conservation Manual. Reef Ranch Publishing Ltd, Leeds, West Yorkshire, UK. https://www.reefranch.co.uk
Austin, B. & Austin, D., A. (2016) Bacterial fish pathogens, 6th ed. Springer, Stirling, UK
Austin, B. (1982) Taxonomy of bacteria isolated from a coastal, marine fish-rearing unit. J. Appl. Bacteriol. 53, 253-268. https://www.doi.org/10.1111/j.1365-2672.1982.tb04684.x
Bass, D., Stentiford. G., D., Wang, H-C., Koskella, B. & Tyler, C., R. (2019) The pathobiome in animal and plant diseases. Trends Ecol Evol. 34(11), 996-1008.
Bell, G., R., Hoskins, G., E. & Hodgkiss, W. (1971) Aspects of the characterization, identification, and ecology of the bacterial flora associated with the surface of stream-incubating Pacific salmon (Oncorhynchus) eggs. J. Fish. Board Can. 28, 1511-1525. https://www.doi.org/10.1139/f71-232
Birrell, C., Mccook, L., Willis, B. & Diaz-Pulido, G. (2008) Effects of benthic algae on the replenishment of corals and the implications for the resilience of coral reefs. Oceanography and marine biology. 46,.
Blazer, V., Phillips, S. & Pendleton, E. (2016) Fish Health, Fungal Infections, and Pfiesteria: The Role of the U.S. Geological Survey. U.S. Geological Survey Fact Sheet. pp 114-198.
Boilard, A., Dubé, C., E., Gruet, C., Mercière, A., Hernandez-Agreda, A. & Derome, N. (2020) Defining Coral Bleaching as a Microbial Dysbiosis within the Coral Holobiont. Microorganisms. 8, 1682. https://doi.org/10.3390/microorganisms8111682
Bone, Q., Marshall, N., B. & Blaxter, J., H., S. (1995) Biology of Fishes. Glasgow: Blackie Academic & Professional. https://www.doi.org/10.1007/978-1-4615-2664-3
Bozzi, D., Rasmussen, J., A., Carøe, C., et al. (2021) Salmon gut microbiota correlates with disease infection status: potential for monitoring health in farmed animals. Anim Microbiome. 3(1), 1-17.
Brandley, B., K. & Schnaar, R., L. (1986) Cell-Surface Carbohydrates in Cell Recognition and Response. J Leukoc Biol 40, 97–111. https://doi.org/10.1002/jlb.40.1.97
Brown, R., Moore, L., Mani, A., Patel, S., Salinas, I. (2021) Effects of ploidy and salmonid alphavirus infection on the skin and gill microbiome of Atlantic salmon (Salmo salar). PLoS One. 16(2), e0243684.
Cámara-Ruiz, M., Cerezo, I., M., Guardiola, F., A. et al. (2021) Alteration of the immune response and the microbiota of the skin during a natural infection by Vibrio harveyi in European seabass (Dicentrarchus labrax). Microorganisms. 9(5), 964.
Carnevali, O., Maradonna, F. & Gioacchini, G. (2017) Integrated control of fish metabolism, wellbeing and reproduction: the role of probiotic. Aquaculture 472, 144-155. https://www.doi.org/10.1016/j.aquaculture.2016.03.037
Cerezuela, R., Meseguer, J. & Esteban, M. (2011) Current knowledge in synbiotic use for fish aquaculture: a review. J. Aquac. Res. Dev. 1, 1-7.
Cherrak, Y., Flaugnatti, N., Durand, E., Journet, L. & Cascales, E. (2019) Structure and Activity of the Type VI Secretion System. Microbiology spectrum, 7(4), 1.
Colorni, A., Avtalion, R., Knibb, W., Berger, E., J., Colorni, B. & Timan, B. (1998) Histopathology of sea bass (Dicentrarchus labrax) experimentally infected with Mycobacterium marinum and treated with streptomycin and garlic (Allium sativum) extract. Aquaculture. 160, 1-17.
Cordero, H., Guardiola, F., A., Tapia-Paniagua, S., T., Cuesta, A., Meseguer, J., Balebona, M., C., et al. (2015) Modulation of immunity and gut microbiota after dietary administration of alginate encapsulated Shewanella putrefaciens Pdp11 to gilthead seabream (Sparus aurata L.). Fish Shellfish Immunol. 45, 608-618. https://www.doi.org/10.1016/j.fsi.2015.05.010
Das, P., Mandal, S., Khan, A., Manna, S., K. & Ghosh, K. (2014) Distribution of extracellular enzyme-producing bacteria in the digestive tracts of 4 brackish water fish species. Turk. J. Zool. 38, 79-88. https://www.doi.org/10.3906/zoo-1205-3
Delcroix, J., Gatesoupe, F., J., Desbruyères, E., Huelvan, C., Le Delliou, H., Le Gall, M., M., Quazuguel, P., et al. (2015) The effects of dietary marine protein hydrolysates on the development of sea bass larvae, Dicentrarchus labrax, and associated microbiota. Aquac. Nutr. 21, 98-104. https://www.doi.org/10.1111/anu.12139
Duperron, S., Halary, S., Habiballah, M., Gallet, A., Huet, H., Duval, C. et al. (2019) Response of fish gut microbiota to toxin-containing cyanobacterial extracts: a microcosm study on the Medaka (Oryzias latipes). Environ SciTechnol Lett. 6, 341–7.
Egerton, S., Culloty, S., Whooley, J., Stanton, C. & Ross, R., P. (2018) The Gut Microbiota of Marine Fish. Frontiers in Microbiology. 9(873),. https://www.frontiersin.org/articles/https://doi.org/10.3389/fmicb.2018.00873/full
Elhassan, Y., Philp, A. & Lavery, G. (2017) Targeting NAD+ in Metabolic Disease: New Insights Into an Old Molecule. Journal of the Endocrine Society. 1(7), 816-835.
Estruch, G., Collado, M., Peñaranda, D., Vidal, A., T., Cerdá, M., J., Martínez, G., P., et al. (2015) Impact of fishmeal replacement in diets for gilthead sea bream (Sparus aurata) on the gastrointestinal microbiota determined by pyrosequencing the 16S rRNA gene. PLoS One 10, e0136389. https://www.doi.org/10.1371/journal.pone.0136389
Flerova, E. & Balabanova, L. (2013) Ultrastructure of granulocytes of teleost fish (Salmoniformes, Cypriniformes, Perciformes). Journal of Evolutionary Biochemistry and Physiology. 49(2), 223-233.
Fournier, V., Huelvan, C. & Desbruyeres, E. (2004) Incorporation of a mixture of plant feedstuffs as substitute for fish meal in diets of juvenile turbot (Psetta maxima). Aquaculture 236, 451-465. https://www.doi.org/10.1016/j.aquaculture.2004. 01.035
Gallet, A., Halary, S., Duval, C., Huet, H., Duperron, S. & Marie, B. (2023) Disruption of fish gut microbiota composition and holobiont’s metabolome during a simulated Microcystis aeruginosa (Cyanobacteria) bloom. Microbiome. 11, 108. https://doi.org/10.1186/s40168-023-01558-2
Ghanbari, M., Kneifel, W. & Domig, K., J. (2015) A new view of the fish gut microbiome: advances from next-generation sequencing. Aquaculture 448, 464-475. https://www.doi.org/10.1016/j.aquaculture.2015.06.033
Giatsis, C., Sipkema, D., Smidt, H. et al. (2015) The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci Rep. 5(1), 1-15.
Green, T., J., Smullen, R. & Barnes, A., C. (2013) Dietary soybean protein concentrate-induced intestinal disorder in marine farmed Atlantic salmon, Salmo salar is associated with alterations in gut microbiota. Vet. Microbiol. 166, 286-292. https://www.doi.org/10.1016/j.vetmic.2013.05.009
Hansen, G. & Olafsen, J. (1999) Bacterial interactions in early life stages of marine cold water fish. Microb. Ecol. 38, 1-26. https://www.doi.org/10.1007/s002489900158
Hlongwane, P., Mungra, N., Madheswaran, S., Akinrinmade, O., A., Chetty, S. & Barth, S. (2018) Human Granzyme B Based Targeted Cytolytic Fusion Proteins. Biomedicines. 6(2), 72.
Ho, J. & Kim, I. (2001) New species of Hatschekia Poche, 1902 (Copepoda: Hatschekiidae) parasitic on marine fishes of Kuwait. Syst Parasitol. 49, 73-79.
Hooper, L., V. & Gordon, J., I. (2001) Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology 11, 1R-10R. https://doi.org/10.1093/glycob/11.2.1R
Hovanec, T. (2019) Dr. How to harness bacteria to cycle your saltwater tank quickly! | MACNA 2019. BrsTV. https://www.youtube.com/watch?v=zDI7sxqC-ss
James, A., G. (1988) Are clupeid microphagists herbivorous or omnivorous? A review of the diets of some commercially important clupeids. S. Afr. J. Mar. Sci. 7, 161-177. https://www.doi.org/10.2989/025776188784379017
Kapoor, B. & Khawna, B. (1993) The potential spectrum of the gut in teleost fishes. Adv. Fish Res. 1, 221-226.
Karachle, P., K. & Stergiou, K., I. (2010) Gut length for several marine fish: relationships with body length and trophic implications. Mar. Biodivers. Rec. 3, e106. https:///www.doi.org/10.1017/S1755267210000904
Kelly, C. & Salinas, I. (2017) Under pressure: Interactions between commensal microbiota and the teleost immune system. Front. Immunol. 8, 1.
Kim, B.,-R., Shin. J., Guevarra, R., B., et al. (2017) Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol. 27(12), 2089-2093.
Kim, J. & Lee, J., L. (2017) Correlation of Total Bacterial and Vibrio spp. Populations between Fish and Water in the Aquaculture System. Frontiers in Marine Science. 4,.
Kiron, V. (2012) Fish immune system and its nutritional modulation for preventive health care. Anim. Feed Sci. Technol. 173, 111-133. https://www.doi.org/10.1016/j.anifeedsci.2011.12.015
Kline, D., I., Kuntz, N., M., Breitbart, M., Knowlton, N. & Rohwer, F. (2006) Role of elevated organic carbon levels and microbial activity in coral mortality. Marine Ecology Progress Series. 314, 119-125.
Langille, M., G., Zaneveld, J., Caporaso, J., G. et al. (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 31(9), 814-821.
Larsen, A., M. (2014) Studies on the Microbiota of Fishes and the Factors Influencing Their Composition. Auburn, AL: Auburn University.
Lasica, A., Ksiazek, M., Madej, M. & Potempa, J. (2017) The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function. Frontiers in Cellular and Infection Microbiology. 7,.
Legrand, T., P., R., A., Catalano, S., R., Wos-Oxley, M., L., Stephens, F., Landos, M., Bansemer, M., S., Stone, D., A., J., Qin, J., G. & Oxley, A., P., A. (2018) The Inner Workings of the Outer Surface: Skin and Gill Microbiota as Indicators of Changing Gut Health in Yellowtail Kingfish. Front. Microbiol. 8:2664. https://www.doi.org/10.3389/fmicb.2017.02664
Lesel, R., De La Noüe, J. & Choubert, G. (1989) Fecal bacterial flora of rainbow trout under antibiotic treatment: effect of the number of pyloric caeca and the lipid content of food. Aquaculture: A Biotechnology in Progress, Vol. 1. De Pauw, N., Jaspers, E., Ackefors, H. & Wilkins, N. (eds.). Bredene: European Aquaculture Society, 592.
Li, P. & Gatlin, D., M. (2003) Evaluation of brewers yeast (Saccharomyces cerevisiae) as a feed supplement for hybrid striped bass (Morone chrysopsX—M. saxatilis). Aquaculture. 219(1), 681-692.
Li, T., Li, H., Gatesoupe, F-J. et al. (2017) Bacterial signatures of ‘red-operculum’ disease in the gut of crucian carp (Carassius auratus). Microb Ecol. 74(3), 510-521.
Liu, Q., Lai, Z., Gao, Y. et al. (2021) Connection between the gut microbiota of largemouth bass (Micropterus salmoides) and microbiota of the pond culture environment. Microorganisms. 9(8), 1770.
Llewellyn, M., Leadbeater, S., Garcia, C. et al. (2017) Parasitism perturbs the mucosal microbiome of Atlantic Salmon. Sci Rep. 7(1), 1-10.
Llewellyn, M., S., Boutin, S., Hoseinifar, S., H. & Derome, N. (2018) Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front Microbiol. 2014, 5. http://journal.frontiersin.org/article/10.3389/fmicb.2014.00207/abstract
Lobo, C., Moreno-Ventas, X., Tapia-Paniagua, S., Rodríguez, C., Moriñigo, M., A. & de La Banda, I., G. (2014) Dietary probiotic supplementation (Shewanella putrefaciens Pdp11) modulates gut microbiota and promotes growth and condition in Senegalese sole larviculture. Fish Physiol. Biochem. 40, 295-309. https://www.doi.org/10.1007/s10695-013-9844-0
Lom, J. & Corliss, J. (1971) Morphogenesis and Cortical Ultrastructure of Brooklynella hostilis, a Dysteriid Ciliate Ectoparasitic on Marine Fishes. The Journal of Eukaryotic Microbiology. 18(2), 261-281.
López Nadal, A., Peggs, D., Wiegertjes, G., F. & Brugman, S. (2018) Exposure to Antibiotics Affects Saponin ImmersionInduced Immune Stimulation and Shift in Microbial Composition in Zebrash Larvae. Front Microbiol 9, 2588. https://doi.org/10.3389/fmicb.2018.02588
López-Dóriga, M., Barnes, A., dos Santos, N. & Ellis, A. (2000) Invasion of fish epithelial cells by Photobacterium damselae subsp. piscicida: evidence for receptor specificity, and effect of capsule and serum. Microbiology. 146(1), 21-30.
Lorgen-Ritchie, M., Clarkson, M., Chalmers, L., Taylor, J., F., Migaud, H. & Martin, S., A. M. (2021) A temporally dynamic gut microbiome in Atlantic salmon during freshwater recirculating aquaculture system (RAS) production and post-seawater transfer. Front. Mar. Sci. 8, 869. https://doi.org/10.3389/fmars.2021.711797
Lorgen-Ritchie, M., Webster, T., U., McMurtrie, J., Bass, D., Tyler, C., R., Rowley, A. & Martin, S., A., M. (2023) Microbiomes in the context of developing sustainable intensified aquaculture. Frontiers in Microbiology. 14, 1200997.
Ma, C., Chen, C., Jia, L., He, X. & Zhang, B. (2019) Comparison of the intestinal microbiota composition and function in healthy and diseased Yunlong Grouper. AMB Express. 9(1), 1-11.
Martins, J. & Vasconcelos, V. (2015) Cyanobactins from cyanobacteria: current genetic and chemical state of knowledge. Mar Drugs. 13, 6910-6946.
Mashoof, S. & Criscitiello, M., F. (2016) Fish Immunoglobulins. Biology. 5(4), 45.
McBride, M., J. & Nakane, D. (2015) Flavobacterium gliding motility and the type IX secretion system. Curr. Opin. Microbiol. 28, 72-77.
Merrifield, D., L. & Rodiles, A. (2015) The fish microbiome and its interactions with mucosal tissues. Mucosal Health in Aquaculture. Peatman, E. (ed.) Academic Press, San Diego, CA. pp 273-295.
Mihalitsis, M. & Bellwood, D. (2017) A morphological and functional basis for maximum prey size in piscivorous fishes. PLoS ONE. 12(9),.
Miyake, S., Ngugi, D. K. & Stingl, U. (2015) Diet strongly influences the gut microbiota of surgeonfishes. Mol. Ecol. 24, 656-672. https://www.doi.org/10.1111/mec.13050
Miyake, S., Soh, M., Azman, M., N., Ngoh, S., Y., Orbán, L., Seedorf, H. (2020) Insights into the microbiome of farmed Asian sea bass (Lates cal-carifer) with symptoms of tenacibaculosis and description of Tenacibaculum singaporense sp. nov. Antonie Van Leeuwenhoek. 113(6), 737-752.
Monroig, Ó., Tocher, D., R. & Navarro, J., C. (2013) Biosynthesis of polyunsaturated fatty acids in marine invertebrates: recent advances in molecular mechanisms. Mar. Drugs 11, 3998-4018. https://www.doi.org/10.3390/md11103998
Moon, D. (2021) Boosting NAD+ to Reverse Aging? Overview of NR and NMN. GeneticLifeHacks.com. https://www.geneticlifehacks.com/nad-reversing-aging-overview-of-nr-and-nmn/
Mouchet, M., A., Bouvier, C., Bouvier, T., Troussellier, M., Escalas, A. & Mouillot, D. (2012) Genetic difference but functional similarity among fish gut bacterial communities through molecular and biochemical fingerprints. FEMS Microbiol. Ecol. 79, 568-580. https://www.doi.org/10.1111/j.1574-6941.2011. 01241.x
Mougin, J. & Joyce, A. (2022) Reviews in Aquaculture. Fish disease prevention via microbial dysbiosis-associated biomarkers in aquaculture. 15, 579-594. https://doi.org/10.1111/raq.12745
Muñoz-Baquero, M., Lorenzo-Rebenaque, L., Garc´ıa-Va´zquez, F., A., Garc´ıa-Pa´rraga, D., Mart´ınez-Priego, L., De Marco-Romero, G., Gala´n-Vendrell, I., D’Auria, G. & Marco-Jime´nez, F. (2023) Unveiling Microbiome Signature In Inner Body Fluids: Comparison Between Wild And Aquarium Small-Spotted Catshark (Scyliorhinus canicular). Frontiers in Marine Science. https://www.doi.org/10.3389/fmars.2023.1151119
Murdoch, C., C. & Rawls, J., F. (2019) Commensal microbiota regulate vertebrate innate immunity-insights from the zebrafish. Front. Immunol. 10:2100. https://doi.org/10.3389/fimmu.2019.02100
Navarrete, P., Espejo, R. T. & Romero, J. (2009) Molecular analysis of microbiota along the digestive tract of juvenile Atlantic salmon (Salmo salar L.). Microb. Ecol. 57, 550-561. https://www.doi.org/10.1007/s00248-008-9448-x
Naya-Català, F., Piazzon, M., C., Calduch-Giner, J. A., Sitjà-Bobadilla, A. & Pérez-Sánchez, J. (2022) Diet and host genetics drive the bacterial and fungal intestinal metatranscriptome of Gilthead Sea bream. Front. Microbiol. 13, 883738. https://doi.org/10.3389/fmicb.2022.883738
Nayak, S., K. (2010) Role of gastrointestinal microbiota in fish. Aquac. Res. 41, 1553-1573. https://www.doi.org/10.1111/j.1365-2109.2010.02546.x
Neuman, C., Hatje, E., Zarkasi, K. Z., Smullen, R., Bowman, J., P. & Katouli, M. (2016) The effect of diet and environmental temperature on the faecal microbiota of farmed Tasmanian Atlantic Salmon (Salmo salar L.). Aquac. Res. 47, 660-672. https://www.doi.org/10.1111/are.12522
Noga, E., J. (2010) Fish Disease. Diagnosis and Treatment. Second Edition. Wiley-Blackwell, John Wiley & Sons Inc. p 139.
Oliva-Teles, A. & Goncalves, P. (2001) Partial replacement of fishmeal by brewers yeast (Saccaromyces cerevisae) in diets for sea bass (Dicentrarchus labrax) juveniles. Aquaculture. 202(3), 269-278.
Orlandi, I., Alberghina, L. & Vai, M. (2020) Nicotinamide, Nicotinamide Riboside and Nicotinic Acid-Emerging Roles in Replicative and Chronological Aging in Yeast. Biomolecules. 10(4), 604.
Ortiz-Estrada, A., M., Gollas-Galván, T., Martínez-Cordova, L., R. & Martínez-Porchas, M. (2019) Predictive functional profiles using metagenomic 16S rRNA data: a novel approach to understanding the microbial ecology of aquaculture systems. Rev Aquacult. 11(1), 234-245.
Perdiguero, P., Martin-Martin, A., Benedicenti, O., Diaz-Rosales, P., Morel, E., Munoz-Atienza, E., Garcia-Flores, M., Simon, R., Soleto, I., Cerutti, A. & Tafalla, C. (2019) Teleost IgD+Ig- B Cells Mount Clonally Expanded and Mildly Mutated Intestinal IgD Responses in the Absence of Lymphoid Follicles. Cell Reports. 29(13), 4223-4235.
Pérez-Pascual, D., Lunazzi, A., Magdelenat, G., Rouy, Z., Roulet, A., Lopez-Roques, C., Larocque, R., Barbeyron, T., Gobet, A., Michel, G., Bernardet, J. & Duchaud, E. (2017) The Complete Genome Sequence of the Fish Pathogen Tenacibaculum maritimum Provides Insights into Virulence Mechanisms. Frontiers in Microbiology. 8,.
Reid, K., M., Patel, S., Robinson, A., J., et al. (2017) Salmonid alphavirus infection causes skin dysbiosis in Atlantic salmon (Salmo salar L.) post-smolts. PLoS One. 12(3):e0172856.
Richardson, L., L., Sekar, R., Myers, J., L., Gantar, M., Voss, J., D., Kaczmarsky, L., Remily, E., R., Boyer, G., L. & Zimba, P., V. (2007) The presence of the cyanobacterial toxin microcystin in black band disease of corals. FEMS Microbiology Letters. 272(2), 182-187.
Riley, M., A. & Wertz, J., E. (2002) Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 84, 357-364.
Ringø, E., Lødemel, J., B., Myklebust, R., Jensen, L., Lund, V., Mayhew, T., M., et al. (2002) The effects of soybean, linseed and marine oils on aerobic gut microbiota of Arctic charr Salvelinus alpinus L. before and after challenge with Aeromonas salmonicida ssp. salmonicida. Aquac. Res. 33, 591-606. https://www.doi.org/10.1046/j.1365-2109.2002.00697.x
Ringø, E., Zhou, Z., Vecino, J., L., G., Wadsworth, S., Romero, J., Krogdahl, Å., et al. (2016) Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac. Nutr. 22, 219-282. https://www.doi.oeg/10.1111/anu.12346
Ritchie, K., B. & Smith, G., W. (1995) Preferential carbon utilization by surface bacterial communities from water mass, normal, and white-band diseased Acropora cervicornis. Mol Mar Biol Biotechnol. 4, 345-354.
Rohde, K. (1932) Ciliophora (ciliates). Marine Parasitology. Rohde, K. (ed.). CSIRO Publishing, Clayton, Australia.
Rombout, J., H., Abelli, L., Picchietti, S., Scapigliati, G. & Kiron, V. (2011) Teleost intestinal immunology. Fish Shellfish Immunol. 31, 616-626. https://www.doi.org/10.1016/j.fsi. 2010.09.001
Rosado, D., Pérez-Losada, M., Pereira, A., Severino, R. & Xavier, R. (2021) Effects of aging on the skin and gill microbiota of farmed seabass and seabream. Anim Microbiome. 3(1), 1-14.
Rosado, D., Pérez-Losada, M., Severino, R. & Xavier, R. (2022) Monitoring infection and antibiotic treatment in the skin microbiota of farmed European seabass (Dicentrarchus Labrax) fingerlings. Microb Ecol. 83(3), 789-797.
Rosado, D., Perez-Losada, M., Severino, R., Cable, J. & Xavier, R. (2019b) Characterization of the skin and gill microbiomes of the farmed seabass (Dicentrarchus labrax) and seabream (Sparus aurata). Aquaculture. 500, 57-64.
Rosado, D., Xavier, R., Severino, R., Tavares, F., Cable, J. & Pérez-Losada, M. (2019a) Effects of disease, antibiotic treatment and recovery trajectory on the microbiome of farmed seabass (Dicentrarchus labrax). Sci Rep. 9(1), 1-11.
Rozas-Serri, M. (2019) Gill diseases in marine salmon aquaculture with an emphasis on amoebic gill disease. CAB Reviews Perspectives in Agriculture Veterinary Science Nutrition and Natural Resources. 14, 1-15.
Santoro, E., P., Borges, R., M., Espinoza, J., L., Freire, M., Messias, C., S., M., A., Villela, H., D., M., Pereira, L., M., Vilela, C., L., S., Rosado, J., G., Cardoso, P., M., Rosado, P., M., Assis, J., M., Duarte, G., A., S., Perna, G., Rosado, A., S., Macrae, A., Dupont, C., L., Nelson, K., E., Sweet, M., J., Voolstra, C., R. & Peixoto, R., S. (2021) Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Science advances. 7(33), 3088.
Santos, F., F., Yamamoto, D., Abe, C., M., Bryant, J., A., Hernandes, R., T., Kitamura, F., C., Castro, F., S., Valiatti, T., B., Piazza, R., Elias, W., P., Henderson, I., R. & Gomes, T. (2019) The Type III Secretion System (T3SS)-Translocon of Atypical Enteropathogenic Escherichia coli (aEPEC) Can Mediate Adherence. Frontiers in microbiology. 10, 1527.
Scapigliati, G., Fausto, A., M. & Picchietti, S. (2018) Fish Lymphocytes: An Evolutionary Equivalent of Mammalian Innate-Like Lymphocytes? Frontiers in immunology. 9, 971.
Segata, N., Izard, J., Waldron, L., et al. (2011) Metagenomic biomarker discovery and explanation. Genome Biol. 12(6), 1-18.
Shabir, U., Ali, S., Magray, A., Ganai, B., Firdous, P., Hassan, T. & Nazir, R. (2018) Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microbial pathogenesis. 114, 50-56.
Sila, A., Nedjar-Arroume, N., Hedhili, K., Chataigné, G., Balti, R., Nasri, M., et al. (2014) Antibacterial peptides from barbel muscle protein hydrolysates: activity against some pathogenic bacteria. LWT Food Sci. Technol. 55, 183-188. https://www.doi.org/10.1016/j.lwt.2013.07.021
Siriyappagouder, P., Galindo-Villegas, J., Dhanasiri, A., K., S., Zhang, Q., Mulero, V., Kiron, V. et al. (2020) Pseudozyma priming influences expression of genes involved in metabolic pathways and immunity in zebrafish larvae. Front. Immunol. 11, 987. https://doi.org/10.3389/fimmu.200.00978
Siriyappagouder, P., Kiron, V., Lokesh, J., Rajeish, M., Kopp, M. & Fernandes, J. (2018) The intestinal mycobiota in wild zebrafish comprises mainly Dothideomycetes while Saccharomycetes predominate in their laboratory-reared counterparts. Front. Microbiol. 9, 387. https://doi.org/10.3389/fmicb.2018.00387
Sitjà-Bobadilla, A., Gil-Solsona, R., Estensoro, I., Piazzon, M., Martos-Sitcha, J., Picard-Sánchez, A., Fuentes, J., Sancho, J., Calduch-Giner, J., Hernández, F. & Pérez-Sánchez, J. (2019) Disruption of gut integrity and permeability contributes to enteritis in a fish-parasite model: a story told from serum metabolomics. Parasites & Vectors. 12,.
Skrodenyte-ArbaCIauskiene, V. (2007) Enzymatic activity of intestinal bacteria in roach Rutilus rutilus L. Fish. Sci. 73, 964-966. https://www.doi.org/10.1111/j.1444-2906.2007. 01421.x
Slinger, J., Adams, M., B., Stratford, C., N., Rigby, M. & Wynne, J., W. (2021) The effect of antimicrobial treatment upon the gill bacteriome of Atlantic salmon (Salmo salar L.) and progression of amoebic gill disease (AGD) in vivo. Microorganisms. 9, 987. https://doi.org/10.3390/microorganisms9050987
Spanova, M. & Daum, G. (2011) Squalene–biochemistry, molecular biology, process biotechnology, and applications. Eur J Lipid Sci Technol. 113(11), 1299-1320.
Spor, A., Koren, O. & Ley, R. (2011) Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279-290. https://doi.org/10.1038/nrmicro2540
Stevens, C., E. & Hume, I., D. (2004) Comparative Physiology of the Vertebrate Digestive System. Cambridge: Cambridge University Press.
Stevens, J., Jackson, R., Olson, J. (2016) Bacteria associated with lionsh (Pterois volitans/miles complex) exhibit antibacterial activity against known fish pathogens. Mar Ecol Prog Ser 558, 167–180. https://doi.org/10.3354/meps11789
Sullam, K., E., Essinger, S., D., Lozupone, C., A., O’Connor, M., P., Rosen, G., L., Knight, R., et al. (2012) Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21, 3363-3378. https://www.doi.org/10.1111/j.1365-294X.2012.05552.x
Swiatecka, D., Markiewicz, L., H. & Wróblewska, B. (2012) Pea protein hydrolysate as a factor modulating the adhesion of bacteria to enterocytes, epithelial proliferation and cytokine secretion–an in vitro study. Cent. Eur. J. Immunol. 37, 227-231. https://www.doi.org/10.5114/ceji.2012.3079
Tapia-Paniagua, S., T., Ceballos-Francisco, D., Balebona, M., C., Esteban, M., A. & Moriñigo, M., A. (2018) Mucus glycosylation, immunity and bacterial microbiota associated to the skin of experimentally ulcered gilthead seabream (Sparus aurata). Fish Shellfish Immunol. 75, 381-390.
Thiéry, R., Cozien, J., Cabon, J., Lamour, F., Baud, M. & Schneemann, A. (2006) Induction of a Protective Immune Response against Viral Nervous Necrosis in the European Sea Bass Dicentrarchus labrax by Using Betanodavirus Virus-Like Particles. Journal of Virology. 80(20), 10201.
Torrecillas, S., Montero, D. & Izquierdo, M. (2014) Improved health and growth of fish fed mannan oligosaccharides: potential mode of action. Fish Shellfish Immunol. 36, 525-544. https://doi.org/10.1016/j.fsi.2013.12.029
Tretina, K., Park, E., S., Maminska, A. & MacMicking, J., D. (2019) Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. The Journal of Experimental Medicine. 216(3), 482-500.
Valero, Y., Saraiva‐Fraga, M., Costas, B. & Guardiola, F., A. (2019) Antimicrobial peptides from fish: beyond the fight against pathogens. Reviews in Aquaculture. 12, 224-253.
Vayssier-Taussat, M., Albina, E., Citti, C., et al. (2014) Shifting the paradigm from pathogens to pathobiome: new concepts in the light of metaomics. Front Cell Infect Microbiol. 4, 29.
Vestrum, R., I., Forberg, T., Luef, B., Bakke, I., Winge, P., Olsen, Y., et al. (2022) Commensal and opportunistic bacteria present in the microbiota in Atlantic cod (Gadus morhua) larvae differentially alter the hosts’ innate immune responses. Microorganisms. 10, 24. https://doi.org/10.3390/microorganisms10010024
Walke, J., B., et al. (2017) Dominance-function relationships in the amphibian skin microbiome. Environ. Microbiol. 19, 3387–3397.
Wang, S. & Loreau, M. (2014) Ecosystem stability in space: α, β and γ variability. Ecol Lett. 17(8), 891-901.
Webster, T., M., U., Rodriguez-Barreto, D., Consuegra, S. & Garcia de Leaniz, C. (2019) Cortisol-induced signatures of stress in the fish microbiome. bioRxiv:826503. https://doi.org/10.1101/826503
Wilson, J., M. & Castro, L., F., C. (2010) Morphological diversity of the gastrointestinal tract in fishes. Fish Physiol. 30, 1-55. https://www.doi.org/10.1016/S1546-5098(10)03001-3
Wilson, M. World Feeds Limited, 3b Coulman Street Industrial Estate, Thorne, Doncaster, DN8 5JS, United Kingdom.
Wu, J., Mao, C., Deng, Y., et al. (2019) Diversity and abundance of antibiotic resistance of bacteria during the seedling period in marine sh cage-culture areas of Hainan, China. Mar Pollut Bull 141:343–349. https://doi.org/10.1016/j.marpolbul.2019.02.069
Xavier, R., Pereira, A., Pagan, A., Hendrick, G., C., Nicholson, M., D., Rosado, D., Soares, M., C., Pérez-Losada, M., Sikkel, P., C. (2020) The effects of environment and ontogeny on the skin microbiome of two Stegastes damselfishes (Pomacentridae) from the eastern Caribbean Sea. Mar Biol. 167(7),1-12.
Xu, Z., Takizawa, F., Casadei, E., Shibasaki, Y., Ding, Y., Sauters, T., J., C., et al. (2020) Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol. 5, 3254. https://doi.org/10.1126/sciimmunol.aay3254
Yatsunenko, T., Rey, F., E., Manary, M., J., Trehan, I., Dominguez-Bello, M., G., Contreras, M., et al. (2012) Human gut microbiome viewed across age and geography. Nature 486, 222-227. https://www.doi.org/10.1038/nature11053
Yoon, J., Matsuo, Y., Matsuda, S., Adachi, K., Kasai, H. & Yokota, A. (2018) Rubritalea sabuli sp. nov., a carotenoid-and squalene-producing member of the family Verrucomicrobiaceae, isolated from marine sediment. Int J Syst Evol Microbiol. 58(4), 992-997.
Yoshimizu, M., Kimura, T. & Sakai, M. (1980) Microflora of the embryo and the fry of salmonids. Bull. Jpn. Soc. Sci. Fish. 46, 967-975. https://www.doi.org/10.2331/suisan.46.967
Yu, Y., Wang, Q., Huang, Z., Ding, L. & Xu, Z. (2020) Immunoglobulins, Mucosal Immunity and Vaccination in Teleost Fish. Frontiers in immunology. 11, 567941.
Zanefeld, J., R. et al. (2017) Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2, 17121.
Zarkasi, K., Z., Taylor, R., S., Abell, G., C., Tamplin, M., L., Glencross, B., D. & Bowman, J., P. (2016) Atlantic salmon (Salmo salar L.) gastrointestinal microbial community dynamics in relation to digesta properties and diet. Microb. Ecol. 71, 589-603. https://www.doi.org/10.1007/s00248-015-0728-y
Zeng, A., Tan, K., Gong, P. et al. (2020) Correlation of microbiota in the gut of fish species and water. 3 Biotech. 0(11), 1-10.
Zhang, L., Ni, C., Xu, W., Dai, T., Yang, D., Wang, Q., Zhang, Y. & Liu, Q. (2016) Intramacrophage Infection Reinforces the Virulence of Edwardsiella tarda. Journal of bacteriology. 198(10), 1534-1542.
Zhang, M., Shan, C., Tan, F., Limbu, S., M., Chen, L. & Du, Z. (2020) Gnotobiotic models: powerful tools for deeply understanding intestinal microbiota-host interactions in aquaculture. Aquaculture 517:734800. https://doi.org/10.1016/j.aquaculture.2019.734800
Zhang, X., Ding, L., Yu, Y. et al. (2018) The change of teleost skin commensal microbiota is associated with skin mucosal transcriptomic responses during parasitic infection by Ichthyophthirius multifillis. Front Immunol. 9, 2972.
Zhang, Y. & Gui, J. (2012) Molecular regulation of interferon antiviral response in fish. Developmental and comparative immunology. 38(2), 193-202.
Zhou, Z., Liu, Y., Shi, P., He, S., Yao, B. & Ringø, E. (2009) Molecular characterization of the autochthonous microbiota in the gastrointestinal tract of adult yellow grouper (Epinephelus awoara) cultured in cages. Aquaculture 286, 184-189. https://www.doi.org/10.1016/j.aquaculture.2008.10.002
Zhou, Z., Yao, B., Romero, J., Waines, P., Ringø, E., Emery, M., et al. (2014) Methodological approaches used to assess fish gastrointestinal communities. Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics, Merrifield, D. & Ringø, E. (eds.). John Wiley & Sons Ltd., Hoboken, NJ.


0 Comments